Figure 4. Orthogonal methods to confirm circular RNA identity.
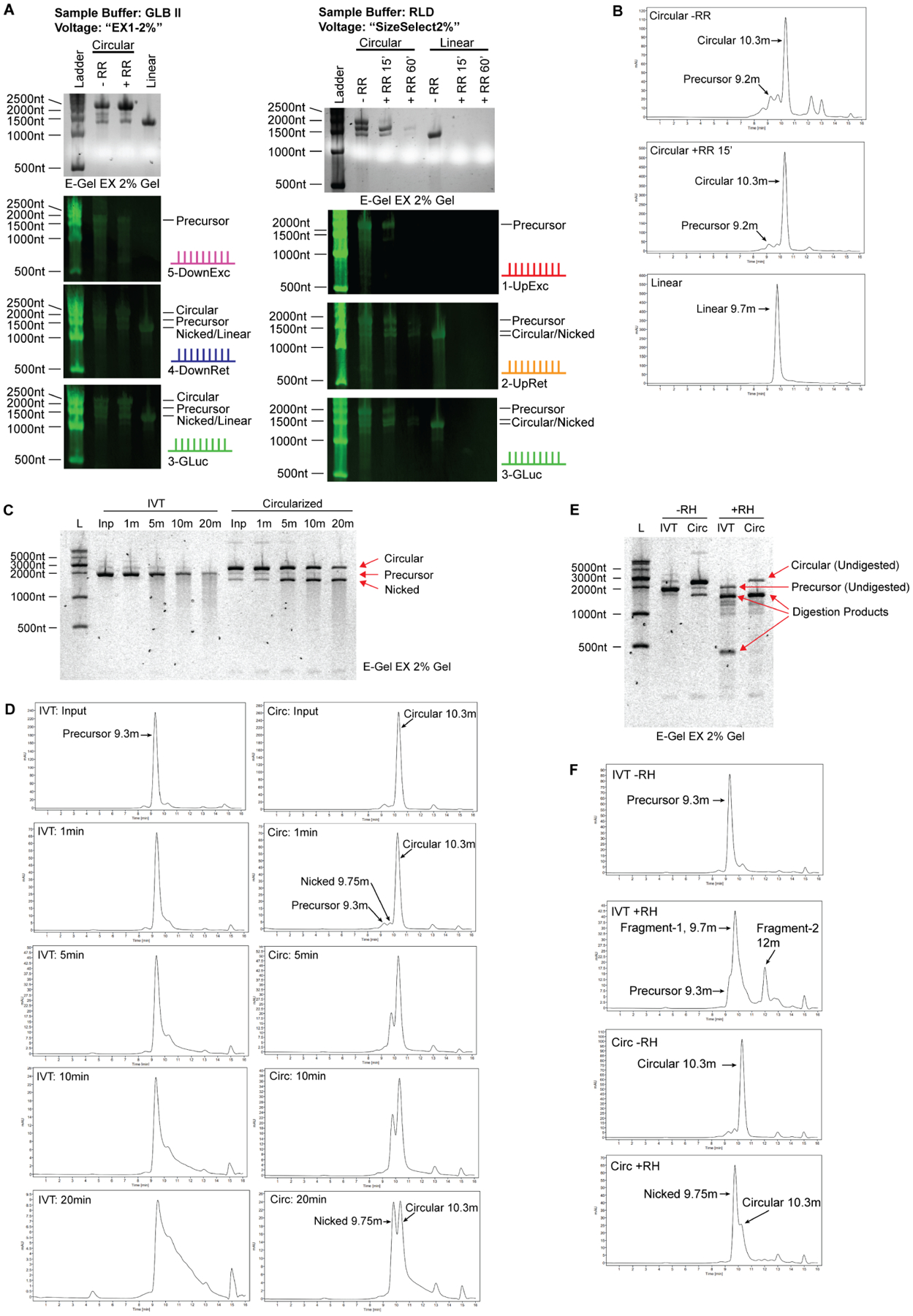
(A) Northern blot analysis of E-Gel EX system. Samples were denatured 1:1 in RLD (2xRNA Loading Dye, ThermoFisher) or GLB II (Gel Loading Buffer II, ThermoFisher), run on E-Gel EX 2% gels and subjected to different voltage programs indicated at the top. Agarose image is shown in top panel, Northern blots shown in lower panels. The probes are depicted on the right of each blot and correspond to the schematic shown in figure 1A.
(B) Size exclusion chromatography of circular RNA. Circular RNA elutes off the column with a slightly smaller molecular weight than predicated. Larger and smaller linear RNA species are on the left and right, respectively. Nicked circular RNA is visible as a minor peak at 9.75 minutes, directly before the major circular RNA peak. RNase R digestion identifies circular RNA as a single enriched peak.
(C) RNA circularity can be validated by nicking the RNA randomly using heat and divalent cations such as Mg2+. 1x T4 RNA ligase I buffer, containing magnesium, was added to RNA sample and RNA in buffer was heated at 70C for the indicated duration. IVT material, which is mostly linear, produces a smear of variable molecular weight species extending from the full-length band when randomly nicked. Circularization reaction material digested with RNase R, which is mostly circular, enriches the lower ‘nicked’ linear RNA band before producing a smear that extends from this band when randomly nicked. No smear extends from the circular RNA band. Note that the top and bottom bands are of equal molecular weight.
(D) Samples in (C) were analyzed by HPLC. Degradation of precursor RNA by heat and magnesium shows typical tailing of RNA towards lower molecular weights, consistent with the smear seen by gel electrophoresis. In contrast, degradation of circular RNA results in enrichment of a new peak corresponding to nicked RNA of equivalent molecular weight, which is followed by tailing as degradation continues. Unlike gel electrophoresis wherein intact circular RNA appears at much higher molecular weight than nicked RNA, circular RNA appears smaller than its nicked counterpart when analyzed using HPLC.
(E) Site-specific degradation using oligonucleotide-guided RNase H digestion is another method for validating RNA circularity. Site specific degradation will yield two linear products when cutting linear RNA species, and one linear product when cutting circular RNA. Predicted degradation products for precursor RNA in this experiment are 1371nt and 399nt in length, visible as the two major product bands in lane 4. Degraded circular RNA yields a single product at 1455nt. Several off-target digestion products are visible as fainter bands.
(F) Samples in (E) were analyzed by HPLC. Degradation of precursor RNA using RNase H yields two smaller fragments. Degradation of circular RNA using RNase H results in enrichment of a single fragment corresponding to nicked RNA of equivalent molecular weight, similar to the product generated by heat/magnesium nicking except that the starts and ends of the nicked molecules are expected to be homogenous.
