Figure 4.
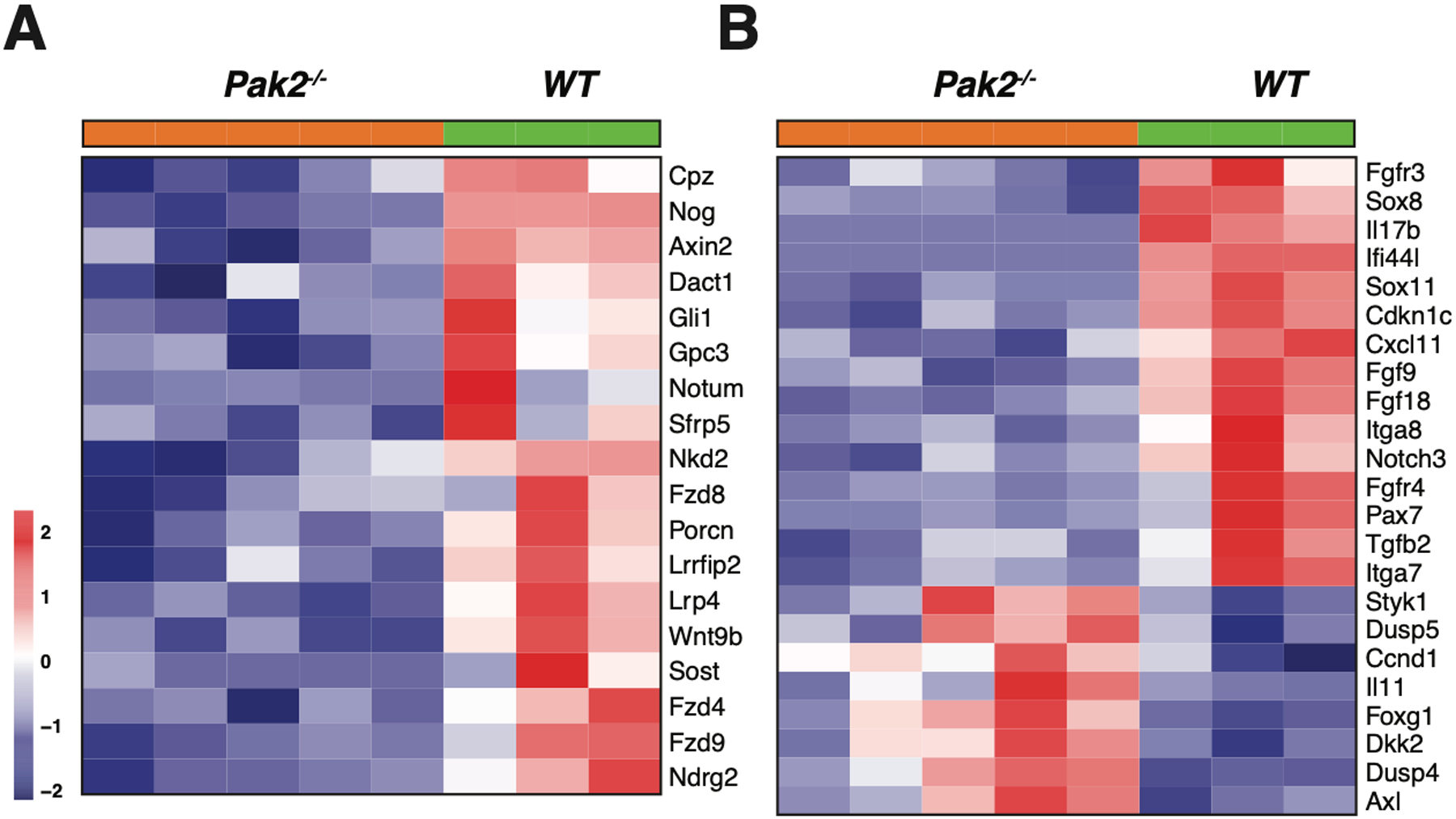

Heatmaps showing expression patterns of genes in NC;P+/+ and NC;P−/− primary malignant mesotheliomas. Heatmaps depict differentially expressed genes observed in peritoneal malignant mesotheliomas from 3 NC;P+/+ mice versus 5 peritoneal malignant mesotheliomas from NC;P−/− mice. (A) Genes involved in Wnt and Hedgehog signaling. (B) Differentially expressed genes involved in multiple pathways/functions, including Akt and Notch signaling, cardiac EMT, cell cycling, cancer stem cell (CSC) markers, and integrins. (C) Validation of a number of differentially expressed genes by semi-quantitative RT-PCR analysis. Left, Down-regulated genes in NC;P−/− primary malignant mesotheliomas include genes involved in Hedgehog signaling (Gli1, Gli2), CSC properties and metastasis (Sox8, Sox11), Wnt signaling (Fzd4, Fzd9, Axin2), and Akt signaling (Igf2, Fgfr4). Right, Validation of genes involved in Notch signaling (Notch3, Hey1) and genes involved in tumor reprogramming in NC;Pak2−/− mesothelioma cells. Note that NC;Pak2−/− cells show downregulation of IfI44L, a tumor suppressor gene that has been implicated in metastasis and drug resistance by regulating met/Src signaling (49), and upregulation of Dkk2 and Styk1, genes involved in cell proliferation and metastasis. Controls include malignant mesothelioma marker genes Msln (mesothelin), Wt1, and cytokeratin 18 (Krt18). Note that in addition to the mutant deleted Pak2 allele seen in NC;P−/− malignant mesotheliomas, there is also a weak wild type allele due to contaminating stroma in these tumor samples. NMC, normal mesothelial cells; *, wild type Pak2 allele; **, mutant Pak2 allele created after adeno-Cre excision of floxed allele. (D) Immunoblot analysis confirming the loss of expression of Pak2 (as well as of Nf2, p16Ink4a and p19Arf) in three NC;P−/− primary malignant mesotheliomas, whereas Pak2 expression is retained in the three NC;P+/+ tumors. NMC, normal mesothelial cells.
