FIGURE 3.
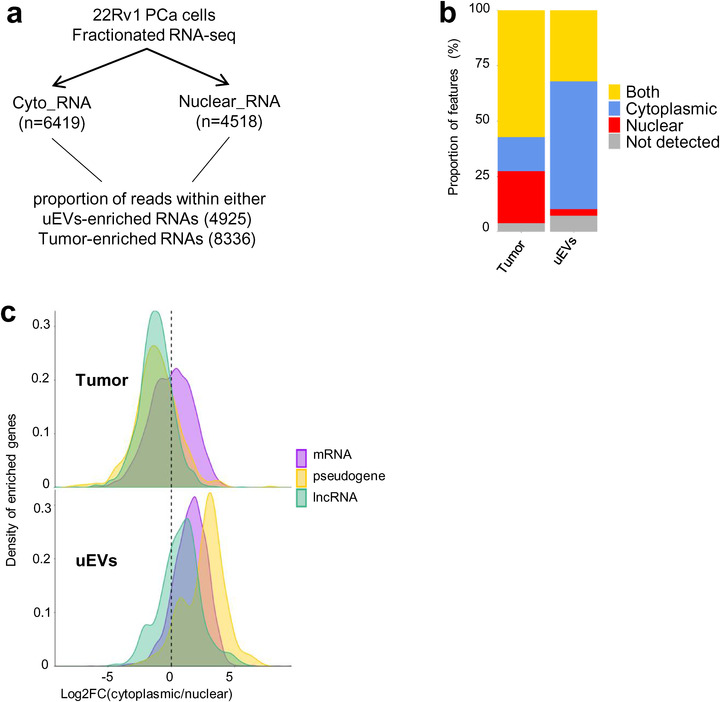
Depleted lncRNAs in uEVs are nuclear. (a). Experimental procedure, starting from 22Rv1 cell line fractionation polyA RNA‐seq, to propose cytoplasmic or nuclear localization of up‐regulated genes in uEVs and up regulated genes in Tumour biopsies. (b). Stacked barplot distribution, by percentage, of cytoplasmic (blue), nuclear (red), both (yellow) or nonpolyA RNAs (grey) of up‐regulated genes in Tumour (8336) and up regulated genes in uEVs (4925). 15.4% and 57.8% upregulated RNAs, respectively, in tumours and uEVs are cytoplasmic; 23.6% and 2.9% are nuclear; 57.2% and 32% are both. (c). Density distribution of log2 (fold change cytoplasmic/nuclear ratio) per RNA types (7732 RNAs from Tumour and 4556 RNAs from uEVs), mRNA (purple), pseudogene (yellow), lncRNA (green) in Tumour (top) and uEVs (bottom). The left side of dotted line in both graphs corresponds to the nuclear RNAs, the right side corresponds to cytoplasmic RNAs
