Figure 2. Mass spectrometry and immunofluorescence of salivary glands from KO mosquitoes.
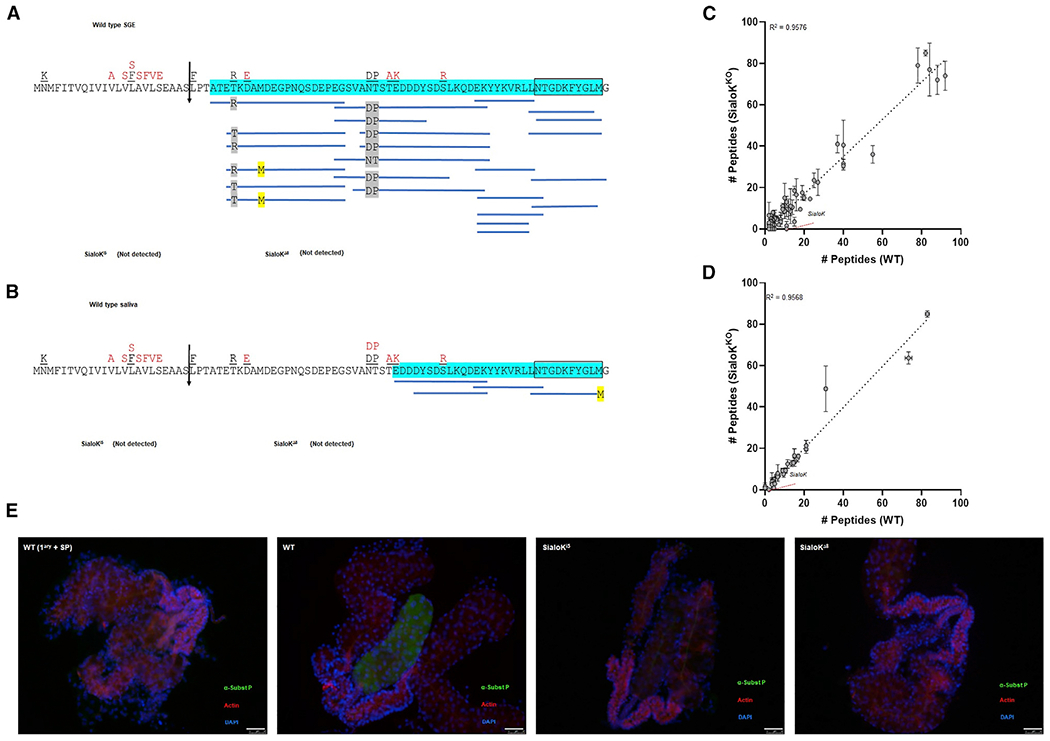
(A and B) Coverage map of sialokinin from (A) SGE obtained from 10 mosquitoes or (B) saliva from 50 mosquitoes by MS/MS. The vertical arrow denotes the predicted secretory cleavage site. Differences in the putative translation product between the cDNA sequence and the genomic sequence are located above the individual amino acids in black (Beerntsen et al., 1999) and red (Ribeiro et al., 2016). A total of 93.55% or 50% of the mature pro-sialokinin sequence was detected by MS/MS (highlighted in blue) in the SGE or saliva, respectively. The active sialokinin decapeptide is boxed. No peptides that matched sialokinin were detected in SGEs or saliva from either sialoKi5 or sialoKΔ8 mosquitoes.
(C and D) Number of unique mapping peptides from SGE (n = 10) or (D) saliva samples from WT and KOs (2 sets of 50 salivating mosquitoes each). For differential expression analysis, the number of unique mapping peptides from sialoKi5 and sialoKΔ8 was combined. Linear regression line and corresponding R2 value are indicated. Bars indicate SDs.
(E) Immunolocalization of sialokinin in the salivary glands of female mosquitoes. A substance P-like protein was not detected in the salivary glands of sialoKi5 and sialoKΔ8 KO mosquitoes. As a negative control (WT 1ary + SP), salivary glands from WT mosquitoes were incubated with anti-substance P primary antibody pre-adsorbed with substance P. Merged fluorescence images are shown in green (anti-substance P; fluorescein isothiocyanate [FITC]), red (phalloidin Alexa Fluor 647), and blue (DAPI). Scale bars, 50 μm.
