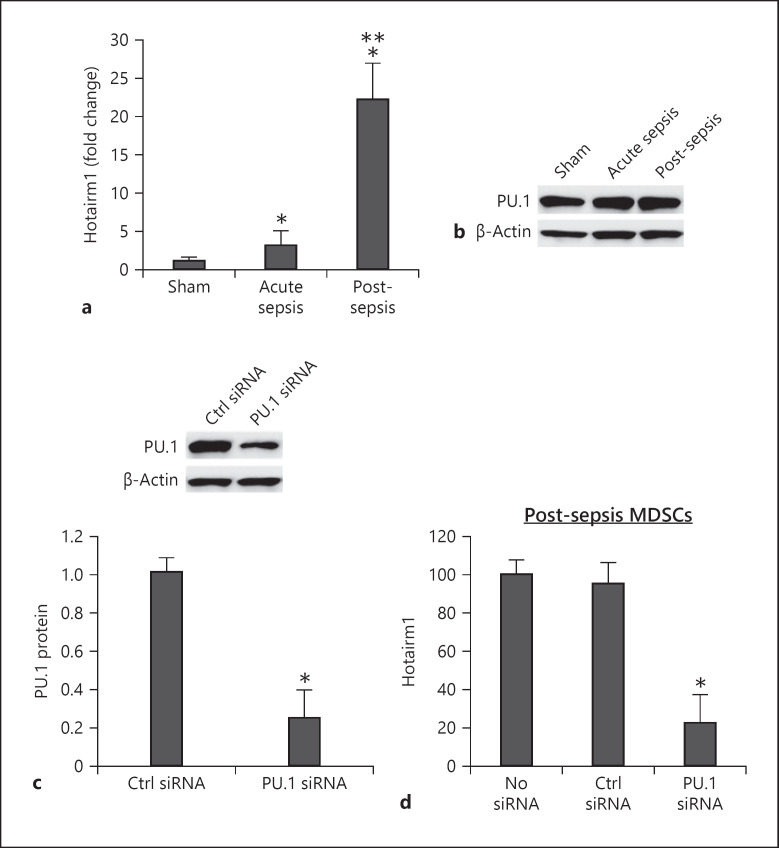
Fig. 2.
Knockdown of PU.1 reduces Hotairm1 expression. Gr1+CD11b+ cells were purified from bone marrow cells by negative selection using magnetic beads and anti-Gr1 and anti-CD11b antibodies. a Total RNA was isolated using TRIzol reagent, and Hotairm1 levels were determined by real-time RT-qPCR using QuantiTect qPCR Primer Assay specific to Hotairm1. Sample values were normalized to GAPDH RNA as an internal control. Data are means ± SD for 5–6 mice per group, from 3 experiments, and are presented relative to sham (1-fold). b Western blot of PU.1 protein in Gr1+CD11b+ cells. The results are representative of 2 blots from 2 experiments. c Knockdown of PU.1 in Gr1+CD11b+ cells. The cells were transfected with PU.1-specific or scrambled/control siRNA for 48 h. PU.1 protein levels were determined by Western blotting. Representative blots and densitometric analysis (lower panel) of 3 blots from 2 experiments are shown. Values were normalized to β-actin and are presented relative to control siRNA. *p < 0.05. d Hotairm1 RNA levels were determined by real-time RT-qPCR as in a. Data are means ± SD for 6 mice per group, from 3 experiments, and are presented relative to nontransfected cells. *p < 0.05 versus no siRNA or control (Ctrl) siRNA. MDSCs, myeloid-derived suppressor cells.