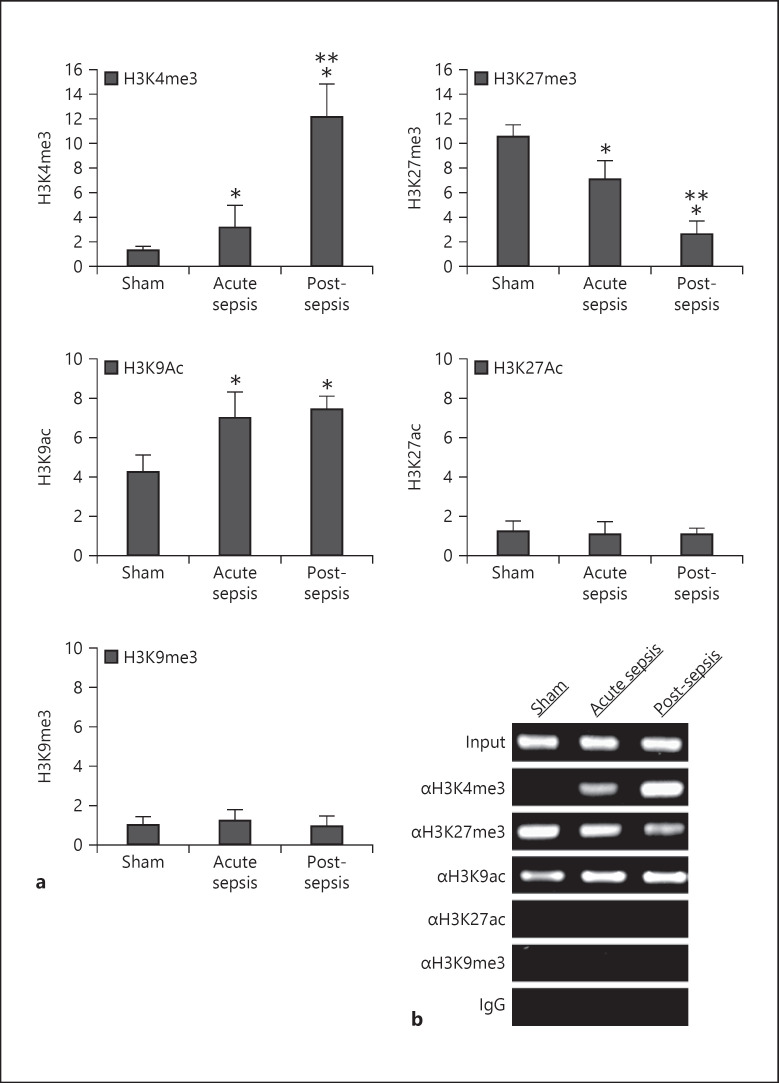
Fig. 4.
Detection of histone modifications at the Hotairm1 proximal promoter. Gr1+CD11b+ cells were purified from bone marrow cells by negative selection using magnetic beads and anti-Gr1 and anti-CD11b antibodies. Chromatin was prepared as described in Figure 1 and immunoprecipitated with antibodies specific to the indicated histone marks or anti-IgG isotype control antibody. a The ChIPed DNA was analyzed by real-time qPCR using primers that amplify the promoter sequences surrounding PU.1 binding site 1. Sample values were normalized to the “input” DNA and are presented as fold change relative to the IgG-immunoprecipitated samples (1-fold). Data are means ± SD for 6–7 mice per group, from 3 experiments. *p < 0.05 versus sham; **p < 0.05 versus acute sepsis. b The ChIPed DNA was amplified by semiquantitative PCR using the same primers as in a. The PCR products (220 bp) were separated by electrophoresis using 1% agarose gel.