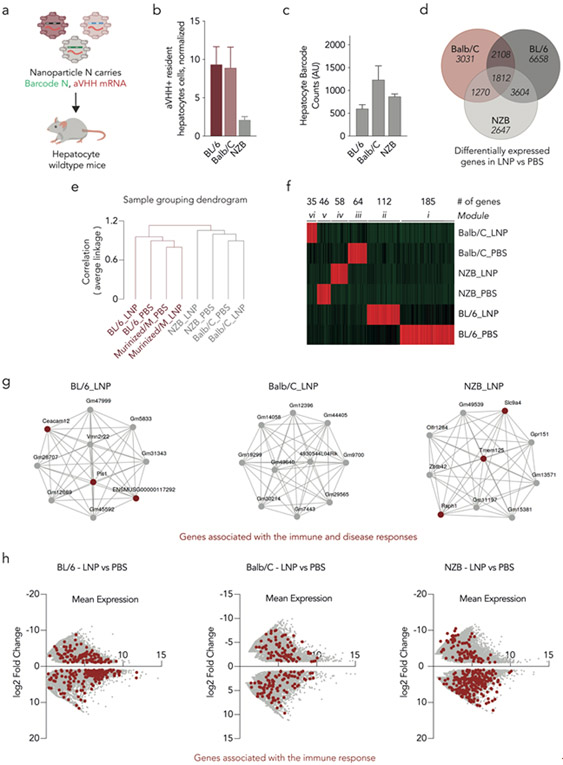
Figure 4.
Inflammatory genes impact mRNA delivery across multiple mouse strains. (a) The 89 LNPs previously described, carrying a DNA barcode and aVHH mRNA, were intravenously administered to wildtype BL/6, Balb/C, and NZB mice. (b) Twenty-four hours later, the percentage of aVHH+ hepatocytes were analyzed. N=3/group, average +/− SEM. (c) Biodistribution of DNA barcodes was quantified using ddPCR. Average +/− SEM. (d) The number of upregulated genes was determined between the different mouse strains, after LNP treatment. (e) Hierarchal clustering analysis was performed indicating different gene expression profiles between Balb/C, NZB, and BL/6. (f) Weighted correlation network analysis (WGCNA) shows the 500 most upregulated genes in wildtype murine hepatocytes; six unique modules were identified. (g) Network analysis from the top 10 upregulated genes found in WGCNA for each cluster of the LNP-treated BL/6 (ii), Balb/C (vi), and NZB (iv). (h) Differential gene expression between the LNP- and PBS-treated samples was analyzed for all three strains. Genes associated with immune response are highlighted in red.