Figure 3. Single cell interrogation of developmental lung injury.
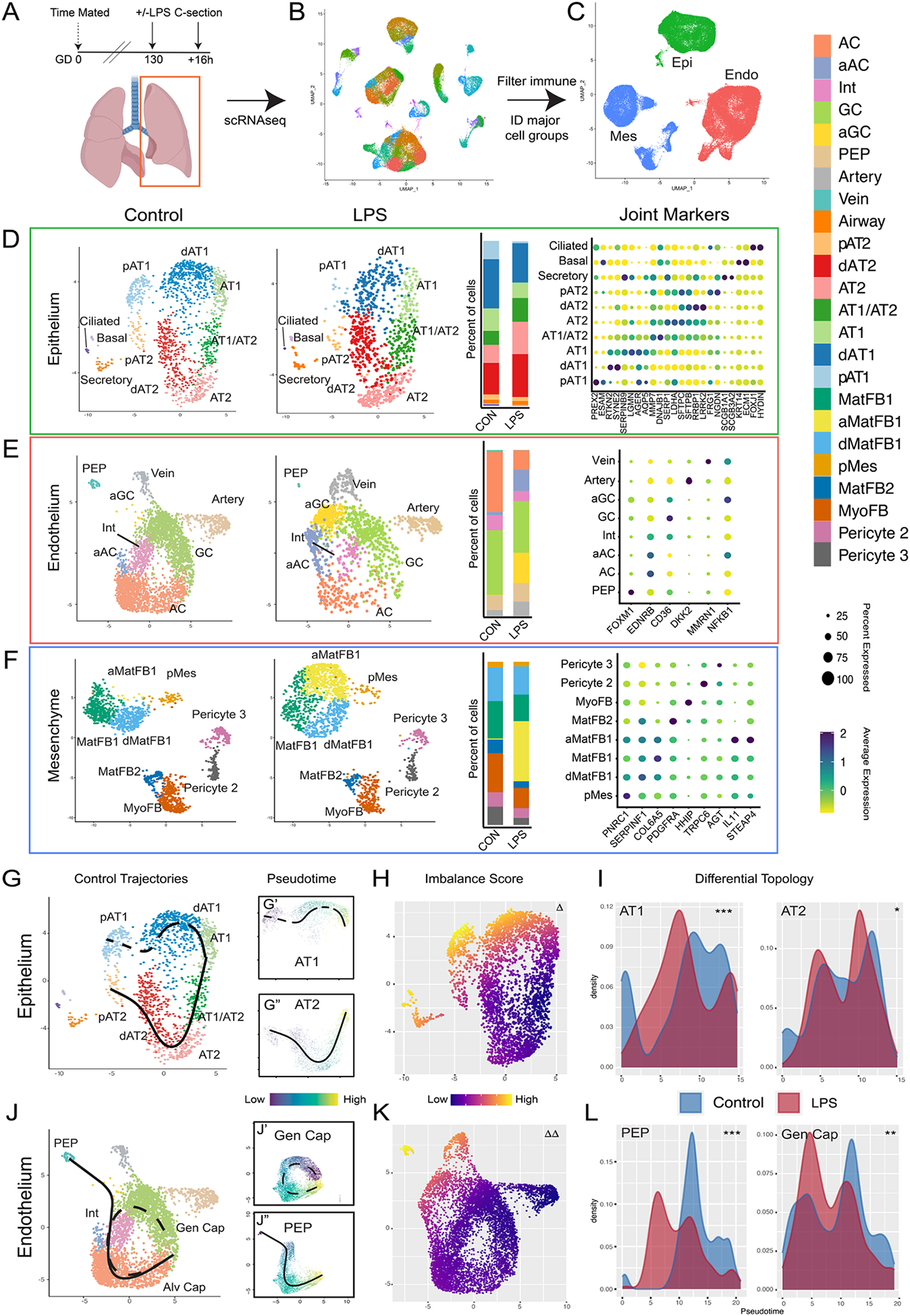
A-C) scRNAseq of control (n=2) and LPS-treated (n=2) primate lungs. UMAP projection of all cells (B) used to identify cells expressing PECAM1/CD31 (Endo), CDH1 (Epi), or COL1A1 (Mes) (C). D-F) Control and LPS UMAPs, cell type proportion per condition, and expression of canonical cell marker genes from epithelial (D), endothelial (E), and mesenchymal (F) cells. G-I) Cellular trajectories from Slingshot show AT1 and AT2 trajectories arising from separable progenitor lineages (G). LPS causes significant transcriptional imbalance (H) in AT1 cells and disrupts development of AT1 but not AT2 cells (I). J-L) Alveolar capillaries arise from both PEP and general capillaries in control lungs (J). LPS disrupts both AC trajectories (K, L). Δ = p <1e-10; Δ Δ = p <1e-13 for differential trajectory topology. *=p<1e-3, **=p<1e-10, ***= p<2.2e-16 for differential trajectory progression. AC = alveolar capillary, aAC = activated AC, Int = intermediate capillary, GC = general capillary, aGC = activated GC, PEP = proliferative endothelial progenitor, MatFB = matrix fibroblast, aMatFB = activated matrix fibroblast, dMatFB = differentiating matrix fibroblast, MyoFB = myofibroblast, pMes = proliferative mesenchyme, AT2 = alveolar type 2, pAT2 = progenitor AT2, dAT2 = differentiating AT2, AT1 = alveolar type 1, pAT1= progenitor AT1, dAT1 = differentiating AT1, CON = control.
