Fig. 4.
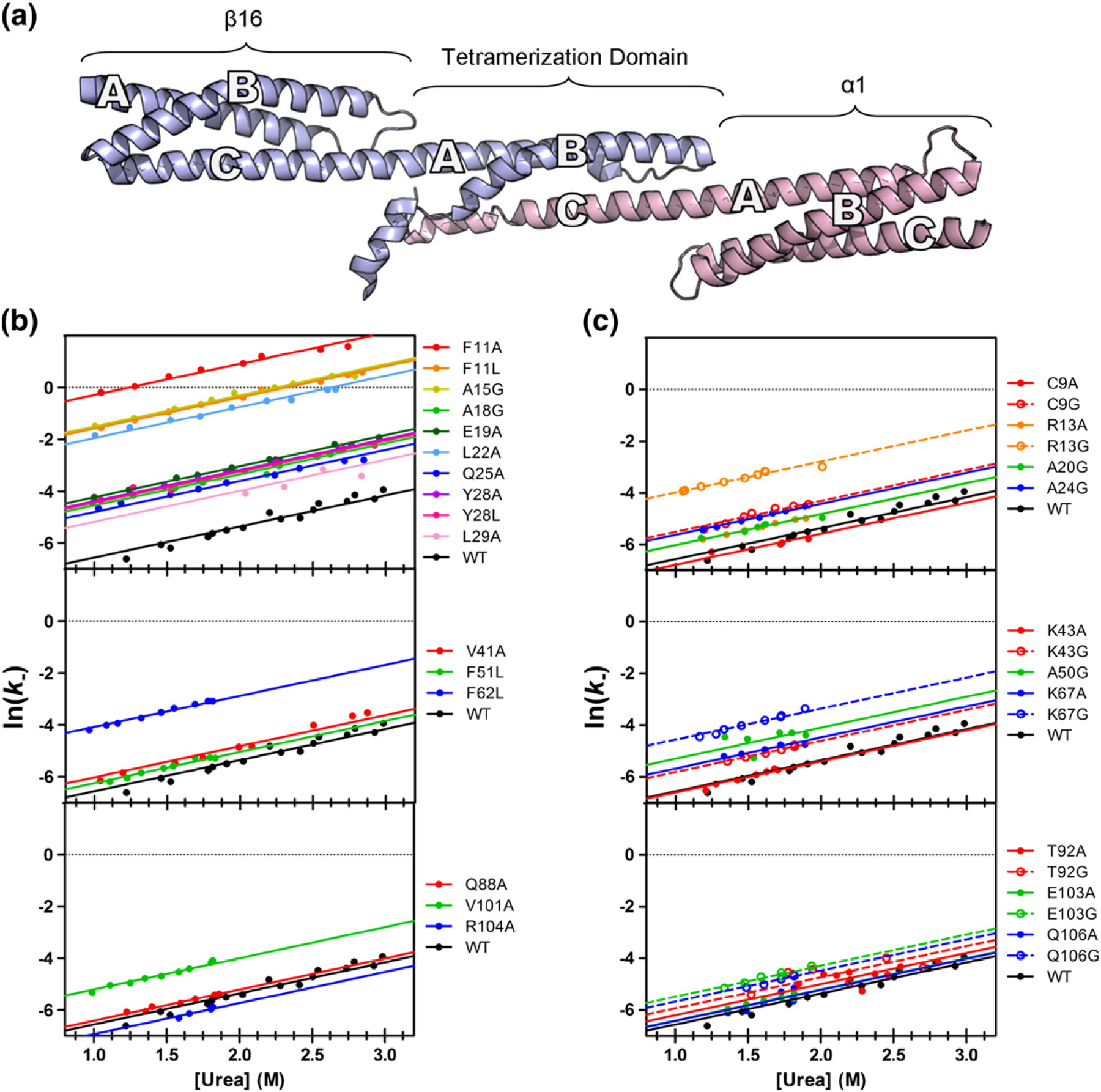
Structure of the tetramer domain and dissociation/unfolding kinetics for mutants. (a) β16β17, shown in blue, consists of a full three-helix domain (β16) followed by a tail (β17), forming the A- and B-helices of the tetramer domain. α0α1 (pink) has a full domain (α1), preceded by the N-terminal tail (α0), which forms the C-helix of the tetramer domain. (b and c) The observed rates for dissociation/unfolding experiments, used to calculate Φ-values for core and surface mutants, respectively. Mutants are divided into three groups: those found on Helix A (top), Helix B (middle), and Helix C (bottom). Each unfolding “limb” is well fitted with an mk− of 1.2 kcal M−1. Mutations to the core of the A-helix show the greatest variation in observed dissociation/folding rate constants.
