Figure 1.
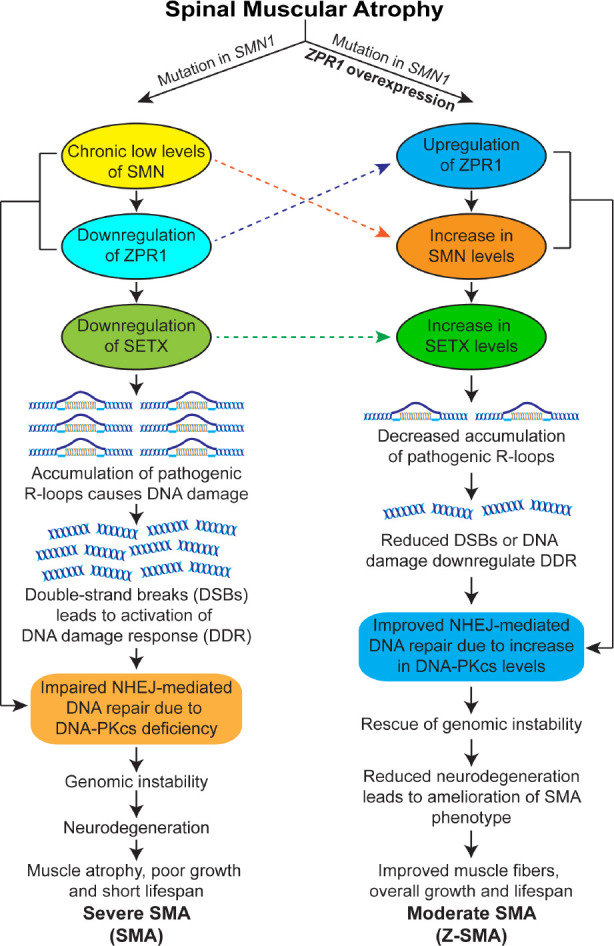
Mechanism of ZPR1-dependent rescue of spinal muscular atrophy disease phenotype.
Mutation in the survival motor neuron 1 (SMN1) gene leads to chronic low levels of the SMN protein. SMN deficiency causes downregulation of ZPR1 and senataxin (SETX), key proteins involved in R-loop resolution. Deficiency of these critical factors results in the accumulation of pathogenic R-loops, causing DSBs and leading to the activation of DNA damage response pathways, genomic instability and neurodegeneration in SMA. Furthermore, the chronic low levels of SMN and ZPR1 result in the deficiency of DNA-dependent protein kinase catalytic subunit (DNA-PKcs), which is critical for non-homologous end joining (NHEJ)-mediated DNA repair. Impairment of NHEJ leads to genomic instability, particularly in neurons (non-proliferating), which rely primarily on NHEJ-mediated DSB repair. R-loop-mediated genomic instability causes degeneration of the spinal cord motor neurons, leading to severe SMA disease with clinical manifestations, including muscle atrophy, reduced growth, and a short life expectancy. ZPR1 overexpression increases the levels of SMN and SETX, leading to decreased accumulation of pathogenic R-loops and rescue of DNA damage in SMA. ZPR1-dependent decrease in R-loop accumulation rescues genomic instability, prevents neurodegeneration, and ameliorates the severity of disease from severe to moderate SMA phenotype. DDR: DNA damage response; DNA-PKcs: DNA-dependent protein kinase catalytic subunit; DSBs: double-stranded breaks; NHEJ: non-homologous end joining; R-loops: RNA-DNA hybrids formed during transcription; SETX: senataxin; SMA: spinal muscular atrophy; SMN: survival motor neuron; Z-SMA: SMA mice with ZPR1 overexpression; ZPR1: zinc finger protein 1.
