Fig. 5 |. Library-scale genome diversification using MAGE-seq and DIvERGE.
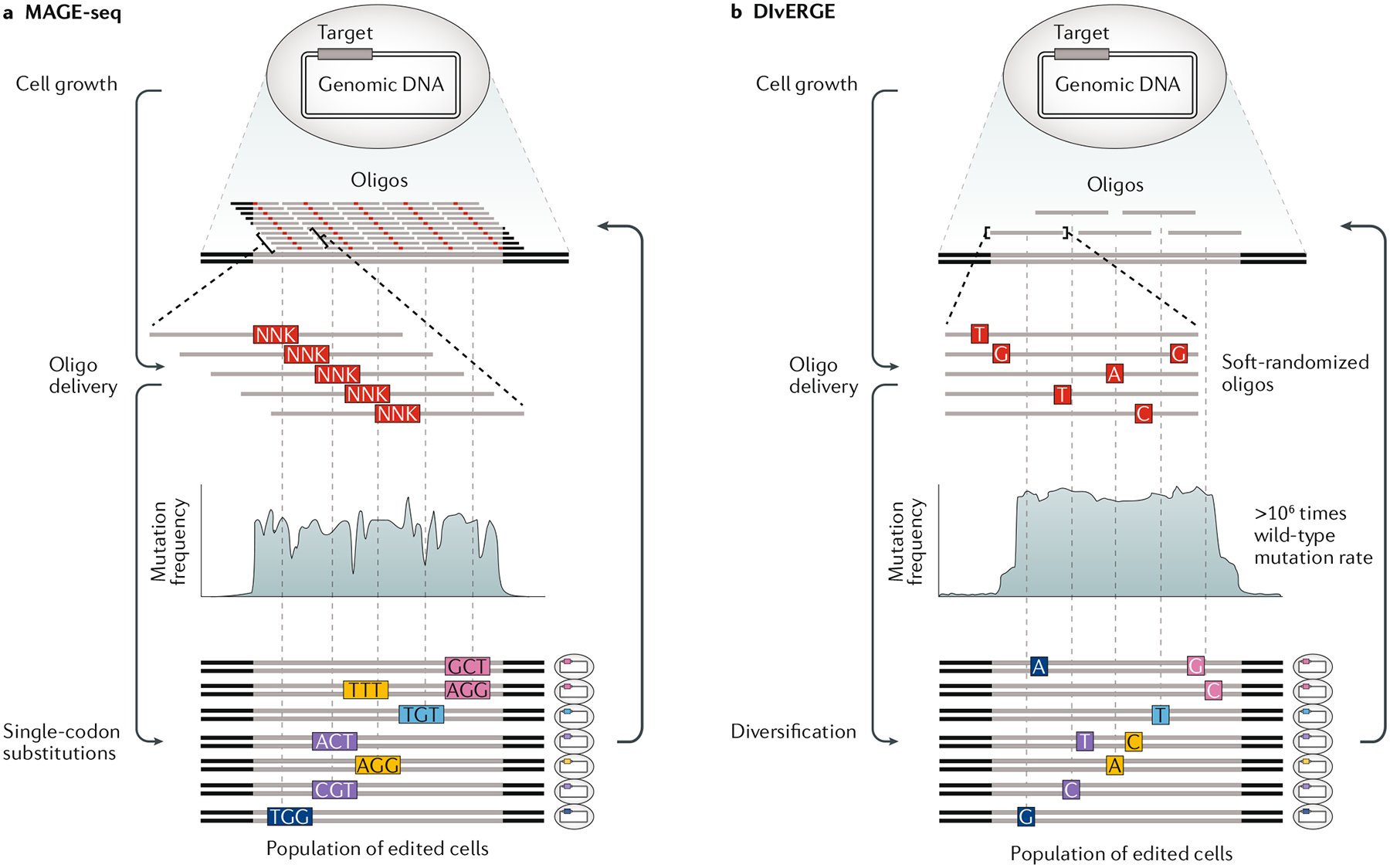
a | Multiplex automated genome engineering with amplicon deep sequencing (MAGE-seq) is based on scanning codon mutagenesis, wherein an oligodeoxynucleotide (oligo) with a degenerate NNK codon is designed for each codon within a targeted gene region. This batch of oligos is pooled and delivered as a library, subjected to multiple MAGE cycles and then a desired phenotype is screened or selected for. The population of enriched cells is then genotyped by amplicon deep sequencing. b | In directed evolution with random genomic mutations (DIvERGE), soft-randomized oligos are designed to tile a targeted genomic locus. As the soft-randomized oligos are incorporated, they will introduce mutations at random, and, depending on the allelic recombination frequency (ARF), this will enable a highly elevated mutation rate to be targeted with precision throughout the genome.
