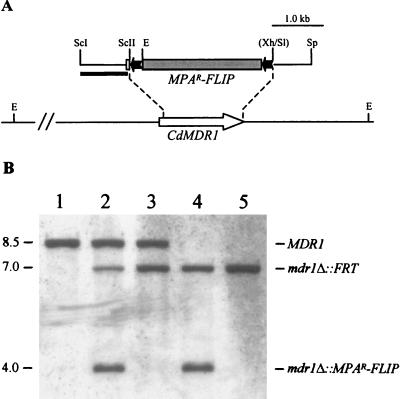
FIG. 1.
Inactivation of the CdMDR1 gene by MPAR flipping. (A) Structure of the CdMDR1 locus in C. dubliniensis strain CM2 and allelic replacements using the insert from pSFIcdM1. Open arrow, CdMDR1 coding region; solid lines, CdMDR1 upstream and downstream sequences. Only relevant restriction sites are shown: E, EcoRI, ScI, SacI, ScII, SacII, Sl, SalI, Sp, SphI, Xh, XhoI. The sites shown in parentheses were destroyed by the cloning procedure. The 5.6-kb MPAR flipper, details of which have been presented elsewhere (21), is not drawn to scale. Solid bar, DNA fragment used as a probe for verification of the correct allelic replacements by Southern hybridization. (B) Southern hybridization of EcoRI-digested genomic DNA of C. dubliniensis parental isolate CM2 and the mdr1 mutant derivatives using the 5′CdMDR1 fragment from pSFIcdM1 as a probe. The identities of the fragments are shown to the right of the blot, and molecular sizes in kilobases are given on the left. Lane 1, CM2 (MDR1/MDR1); lane 2, CdM1 (MDR1/mdr1Δ::MPAR-FLIP); lane 3, CdM2 (MDR1/mdr1Δ::FRT); lane 4, CdM3 (mdr1Δ::MPAR-FLIP/mdr1Δ::FRT); lane 5, CdM4 (mdr1Δ::FRT/mdr1Δ::FRT).