Figure 8.
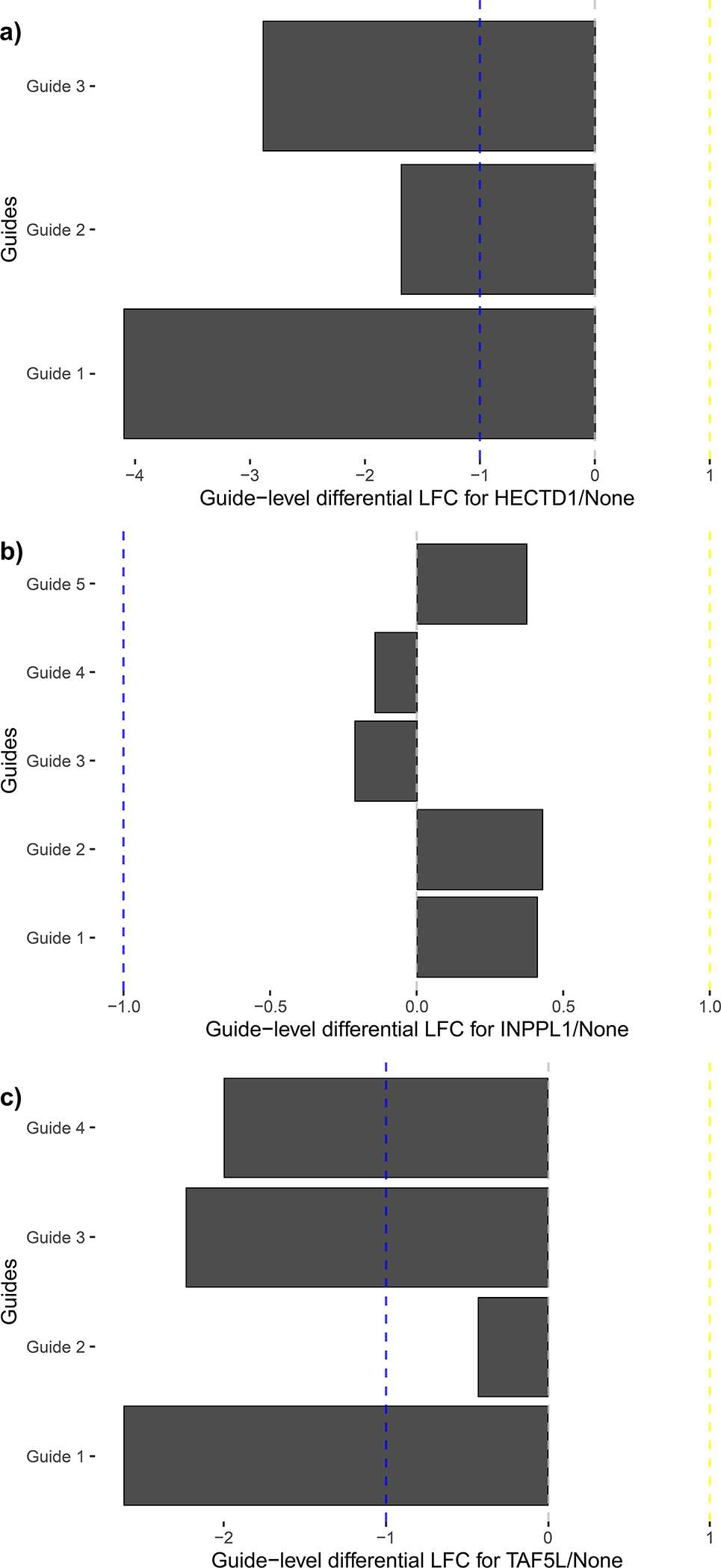
Differential LFC for WT HAP1 guides from the ChyMErA dataset analyzed in Procedure 2 comprising three significant hits of the scored dual-targeting guides at T18. Genes were scored based on deviations from a loess-corrected null model, so the agreement of individual guides of interesting hits should be examined to qualitatively confirm hit quality. (a) is the top-ranked negative hit, HECTD1, with strong agreement for all three guides. (b) is the fourth-ranked negative hit, INPPL1, whose guides indicate a lower hit quality due to three out of five guides possessing positive residual LFCs. (c) is the eighth-ranked negative hit TAF5L, with strong agreement for three guides and no phenotype for the fourth.
