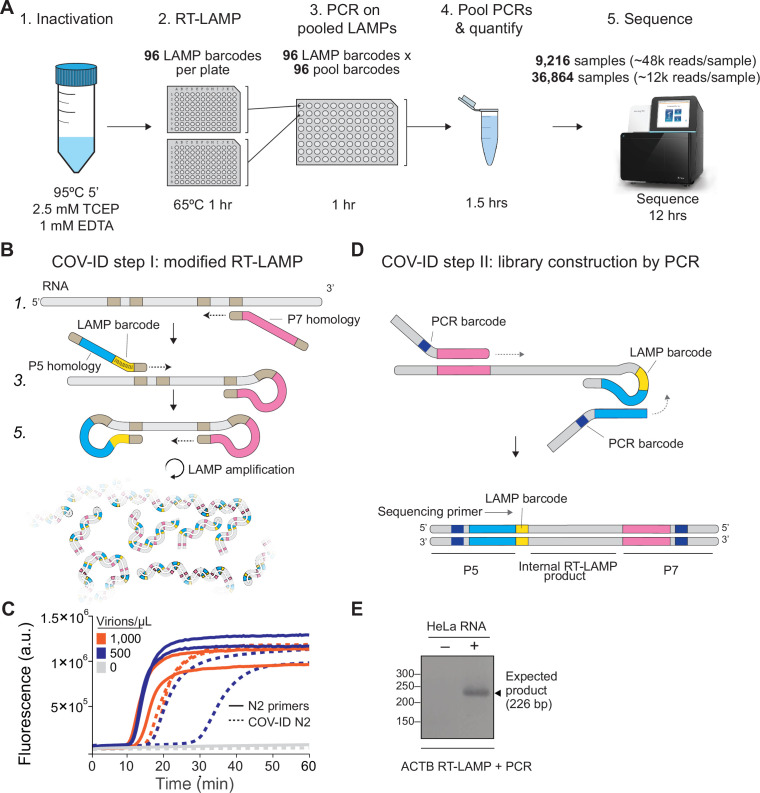
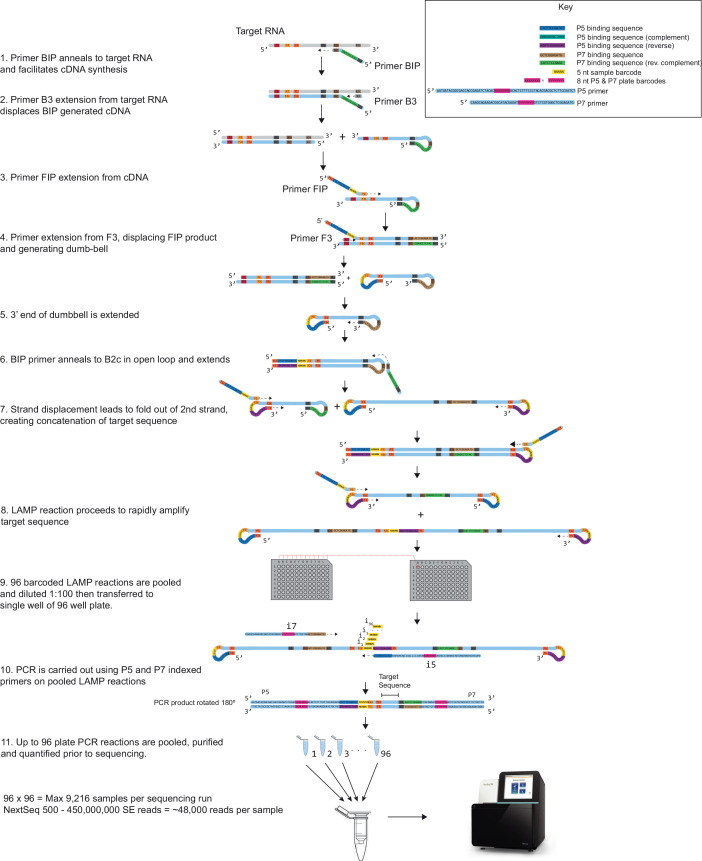
Figure 1. Barcoding and PCR amplification of reverse transcription loop-mediated isothermal amplification (RT-LAMP) products.
(A) Overview of COV-ID. Saliva is collected and inactivated prior to RT-LAMP performed with up to 96 individual sample barcoded primers. LAMP reactions are pooled and further amplified via PCR to introduce Illumina adapter sequences and pool-level dual indexes. A single thermal cycler can amplify 96 or 384 such pools and the resulting ‘super-pool’ can be sequenced overnight to detect multiple amplicons from 9,216 or 36,864 individual patient samples (number of reads in parenthesis assume an output of ~450 M reads from a NextSeq 500). (B) Schematic of the RT-LAMP (step I) of COV-ID. Selected numbered intermediates of RT-LAMP reaction are shown to illustrate how the LAMP barcode, shown in yellow, and the P5 and P7 homology sequences (blue and pink, respectively) are introduced in the final LAMP product. Upon generating the dumbbell intermediate, the reaction proceeds through rapid primed and self-primed extensions to form a mixture of various DNA amplicons containing sequences for PCR amplification. A more detailed version of the LAMP phase of COV-ID, including specific sequences, is illustrated in Figure 1—figure supplement 1. (C) Conventional RT-LAMP primers (solid lines) or primers modified for COV-ID (dotted lines) were used for RT-LAMP of SARS-CoV-2 in saliva. The numbers of inactivated SARS-CoV-2 virions per µL is indicated in the color legend. Each line represents an independent biological replicate. Fluorescence is shown in arbitrary units. (D) Schematic of the PCR (step II) of COV-ID. Following RT-LAMP, up to 96 reactions are pooled and purified and Illumina libraries are generated directly by PCR with dual-indexed P5 and P7 adapters in preparation for sequencing. (E) COV-ID primers targeting ACTB mRNA were used for RT-LAMP from HeLa total RNA. LAMP was diluted 1:100, amplified via PCR and resolved on 2% agarose gel.