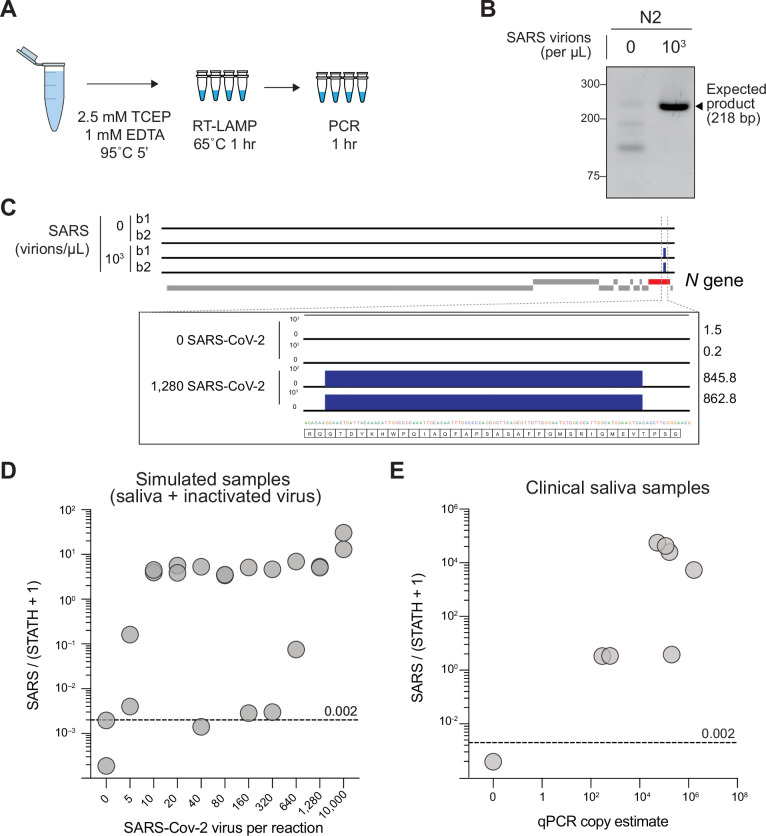
Figure 2. Sequencing-based detection of SARS-CoV-2 in saliva samples.
(A) Saliva preparation. Crude saliva was inactivated via TCEP/EDTA addition and 95 °C incubation prior to RT-LAMP. (B) RT-LAMP followed by COV-ID PCR performed directly on saliva. Saliva with and without addition of 1,000 copies of inactivated SARS-CoV-2 templates was inactivated as described in (A), then used as template. (C) Alignment of sequenced reads against SARS-CoV-2 genome from COV-ID of inactivated saliva spiked with or without 1,280 virions SARS-CoV-2 per µL. All SARS-CoV-2 reads align exclusively to expected region of the N gene. Open reading frames of viral genome are depicted via gray boxes below alignment. Inset: scale shows reads per 1,000. Height of peak is provided on the right. (D) Scatter plot for the ratio of SARS-CoV-2 / (STATH +1) reads obtained by COV-ID (y axis) versus the number of virions per µL spiked in human saliva (x axis). The threshold was set above the highest values scored in a negative control (dashed line). Each circle represents an independent biological replicate. (E) COV-ID performed on clinical saliva samples. The scatter plot shows the SARS-CoV-2 / (STATH +1) read ratio (y axis) versus the viral load in the sample estimated by a clinically approved, qPCR-based diagnostic test. The threshold was set based on the negative controls shown in (D). Each circle represents an independent biological replicate.