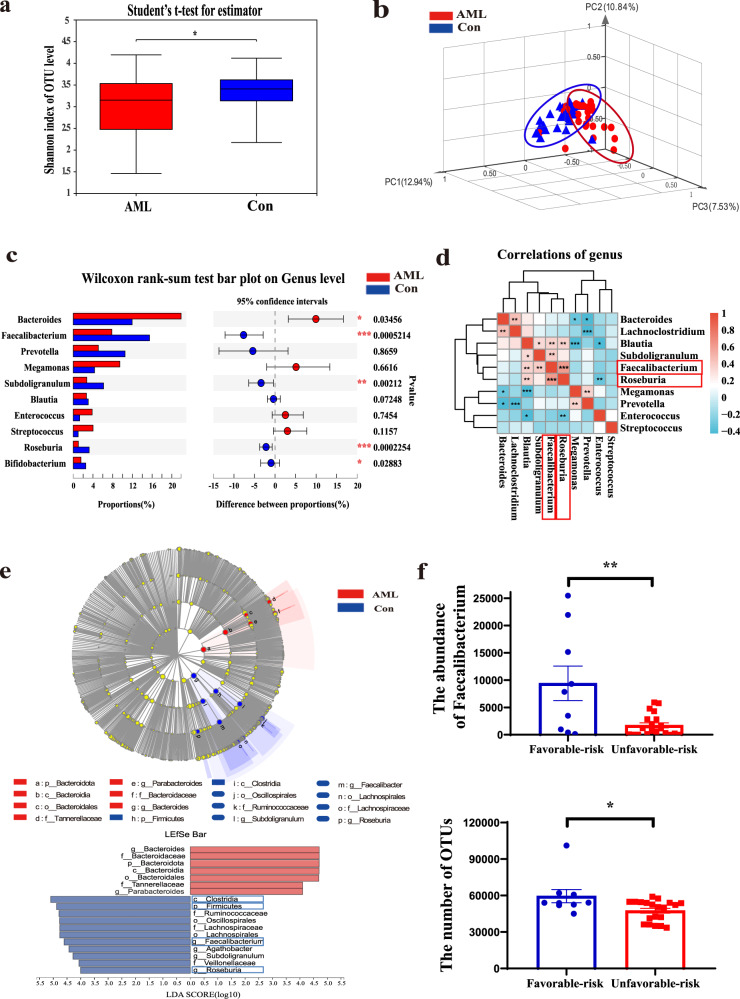
Fig. 1. The diversity and composition of the gut microbiota are significantly altered in AML patients.
Total bacterial DNA was isolated from the intestinal content, and 16S rRNA genes were sequenced. a The diversity and richness of the gut microbiota in AML patients (AML) and healthy controls (Con). Unpaired t-tests were used to compare the Shannon index (n = 61). Data were presented as standard boxplots (with the box encompassing Q1–Q3, the median denoted as a central horizontal line in the box, and the whiskers covering the data within ±1.5 IQR). b PCoA of a weighted UniFrac distance analysis (n = 61). c Relative taxon abundance comparison among the AML and control groups (n = 61). d Spearman correlations between the intestinal content of the 10 genera in AML patients and healthy controls. Faecalibacterium and Roseburia were significantly correlated (red positive correlation, blue negative correlation). e Cladogram generated from linear discriminant analysis effect size (LEfSe) and the LDA score. f The abundance of Faecalibacterium and OTUs was reduced in the unfavourable-risk group (n = 20) compared with the favourable-risk group (n = 9). P values were determined using two-tailed t-test in (a, f) and using Wilcoxon rank test in c. Error bars represent mean ± SEM in (a, c, f). *P = 0.02178 (a), **P = 0.0016, *P = 0.015 (f). Source data are provided as a Source Data file.