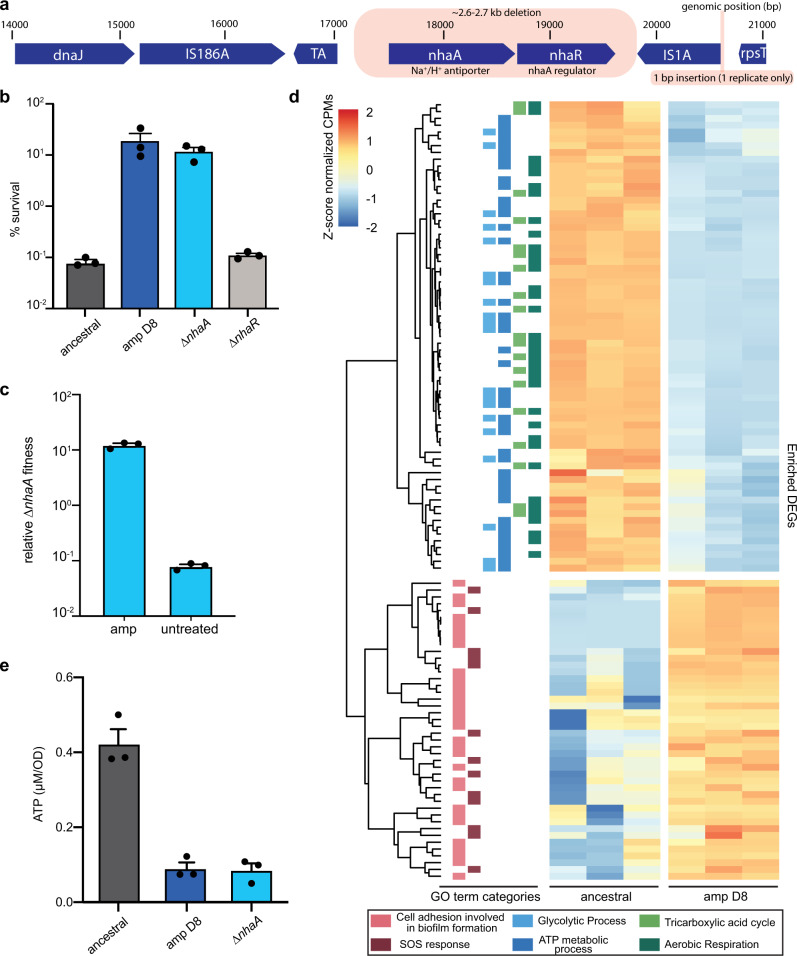
Fig. 2. A novel mechanism of tolerance through nhaA loss.
a Deletion of an ~2.6–2.7 kb region from the E. coli BW25113 genome, including the sodium-proton antiporter nhaA and its regulator nhaR, in all replicates of ampicillin-evolved cells was identified through whole-genome sequencing. One replicate also had a 1 bp insertion near the IS1A insertion element. b Percent survival of ancestral, amp D8, ΔnhaA, and ΔnhaR under ampicillin treatment. Overnight cultures were diluted 1:100 and treated with ampicillin for 6 hours. Shown is the mean of three biological replicates; error bars represent SEM. c Relative ΔnhaA fitness under ampicillin treatment and an untreated control. Overnight cultures of ancestral and ΔnhaA were combined and diluted 1:100 into fresh LB, then treated with ampicillin or a vehicle control (water) for 6 hours. The mean of three biological replicates is shown; error bars indicate SEM. d Hierarchical clustering of differentially expressed genes (DEGs) for three biological replicates of ancestral and amp D8. Heat map color shows z-score normalized counts per million (CPM). DEGs belonging to each GO term category are denoted on the left and in the bottom legend. GO term categories included are cell adhesion involved in biofilm formation (GO:0043708), SOS response (GO:0009432), glycolytic process (GO:0006096), ATP metabolic process (GO:0046034), tricarboxylic acid cycle (GO:0006099), and aerobic respiration (GO:0009060). e Intracellular ATP concentration in ancestral, amp D8, and ΔnhaA. Data are representative of three biological replicates; error bars denote SEM. Source data are provided as a Source Data file.