Abstract
Influenza viruses are simultaneously supported and antagonized by factors within the host cell. This close relationship is the theoretical basis for future antivirals that target the host rather than the virus itself, a concept termed host-directed therapeutics. Genetic screening has led to the identification of host factors capable of modulating influenza virus infections, and these factors represent candidate targets for host-directed antiviral strategies. Despite advances in understanding host targets however, there are currently no host-directed interventions for influenza viruses in clinical use. In this brief review, we discuss some host factors identified in knockout/knockdown and overexpression screens that could potentially be targeted as host-directed influenza intervention strategies. We further comment on the feasibility of changing gene expression in the respiratory tract with RNA delivery vectors and transient CRISPR-mediated gene targeting.
Graphical Abstract
Introduction
Influenza viruses circulate globally resulting in approximately five million hospitalizations and nearly 400,000 deaths annually [1,2]. Though vaccinations are central to limiting disease burden, vaccine hesitancy and suboptimal vaccine efficacy lead to incomplete protection of the population [3]. Thus, even with annual vaccination campaigns, prophylactic and therapeutic antiviral intervention strategies are needed. All currently FDA-approved antivirals that target influenza viruses are small molecules designed to inhibit viral protein functionality [4]. These direct-acting influenza virus antivirals exert a strong selective pressure on the virus, frequently resulting in the fixation of drug-resistant mutations in the viral population [5]. An alternative strategy to prevent or treat viral infections is the generation of host-directed antivirals. Influenza viruses, like all viruses, are obligate intracellular parasites that depend on host cell machinery for replication. In addition to the virus co-opting host proteins to facilitate viral replication, influenza viruses can also be antagonized by cellular innate immune proteins. Host-directed antivirals are based on these concepts and attempt to inhibit/downregulate host dependency factors and/or upregulate viral restriction factors to suppress viral replication. In theory, there are numerous advantages to targeting host factors as opposed to traditional direct-acting antivirals, including a lower probability for the emergence of viral escape mutants and potential for broadly acting action against multiple strains or families of viruses [6].
Despite these advantages, the implementation of host-directed antivirals for clinical use is challenging. Successful host-directed intervention would require the identification of required or restrictive genetic regulators of viral infection as well as the ability to safely modulate the functionality or expression of these targets in a patient. The advent of high-throughput screening strategies has identified hundreds of factors associated with influenza virus infections over the last decade [7–18]. The major limitations toward implementing these genetic hits as intervention strategies, however, have been understanding which of these host factors should be prioritized as candidates as well as developing approaches to modulate the activity of these factors in vivo. Here, we discuss some promising host factor targets discovered via high-throughput screening approaches, evaluate their potentials as host-directed antivirals against influenza virus infection, and explore how gene modulation in a clinical setting could potentially be achieved.
Candidate host-factor targets for host-directed intervention strategies
Evaluation of host factors from genetic screens
The interactions between influenza viruses and host cell factors have been extensively studied by numerous high-throughput methods in a wide variety of experimental systems [7–18]. Additionally, results from these screens have been further compared computationally to identify recurring hits in several meta-analysis “omics” studies [18–20]. For the purposes of this review, we endeavored to highlight a brief list of factors that may be highly promising for the development of clinical interventions. To assess the potential of a host factor for host-directed intervention strategies, we considered four overarching questions with respect to the host factors reported in the studies highlighted above: (1) Has the factor been reported or tested in independent studies? (2) Is the mechanism by which the host factor promotes/restricts infection generally well-understood? (3) Is the host factor relevant/predicted to be relevant to infections in vivo? (4) Has the host factor been safely modulated in vivo or is there an anticipated threat of toxicity to the host? We will focus on four major groups of host factors that have high potential for the development of these therapeutics: Modifiers of the viral receptor, controllers of viral endocytosis, ubiquitin ligases, and interferon-stimulated genes. The pros and cons of targeting the specific host factors within these groups are described in detail in the following sections.
Cell surface expression of the sialic acid receptor
One of the most reproducible strategies for the reduction of influenza virus infection in vitro is limiting the availability of sialic acid on the surface of cells. Sialic acid serves as the cellular receptor for the entry of all influenza viruses that circulate in humans, and sialic acid linkage orientation determines the tropism of infection in the respiratory tract [21]. Several host factors are required for sialic acid biosynthesis and expression on the cell surface [22], however total depletion is most easily achieved in vitro by genetic knockout of the genes encoding sialic acid transporters SLC35A1 and/or SLC35A2. When these transporters are lost, the sugar is thought to be unavailable to glycosyltransferases in the secretory pathway [17]. Interestingly, either SLC35A1 or SLC35A2 was identified as a host-dependency factor by all haploid and CRISPR knockout screens with influenza virus [8,17,18], but only identified as a significant hit in one siRNA screen [9]. However, the demonstration of successful downregulation of SLC35A1/2 in vivo has not occurred, likely because sialylation is required for murine embryonic development which precludes testing in complete knockout models [23].
Access to sialic acid by influenza virions can also be blocked sterically with the addition of sugars to canonical glycans on epithelial cells. We previously reported that upregulation of the glycosyltransferase B4GALNT2 restricted influenza viruses that are tropic for α2,3 linked sialic acid [16]. Restriction is mediated by the addition of a GalNAc onto the subterminal galactose of N-linked glycans with terminal sialic acid moieties. Addition of GalNAc to the subterminal galactose was sufficient to prevent binding of diverse avian influenza viruses to cells. This strategy has further been independently validated in porcine cell systems demonstrating the opportunity to reduce circulation of avian influenza strains in livestock [24]. However, it remains to be determined if upregulation of B4GALNT2 in vivo would be efficacious and tolerated by the host.
For both elimination and/or modification of sialic acid in the lungs, there are potential challenges regarding clinical tolerance. Sialic acid has also been shown to create an anionic barrier in the lungs which may promote airway hydration and provide protection to the epithelium [25]. Despite this, application of a bacterial sialidase to the respiratory tract removes sialic acid moieties without observable toxicity in mice and early clinical data has suggested that sialidase treatment in human patients is generally well-tolerated [26,27]. Transient modulation of glycan transporters or modifying enzymes in the respiratory tract may therefore represent a promising prospective candidate for host-directed prophylactic approaches.
Endosomal acidification machinery
After binding to sialic acid, influenza viruses enter host cells via receptor-mediated endocytosis [28]. Acidification of the resulting endosome is required for the release of the viral genome to the cytoplasm. Accordingly, functional subunits of the vacuolar-type ATPase (V-ATPase) and factors required for V-ATPase endosomal assembly were repeatedly identified in high-throughput screening experiments [18,29]. In the absence of the functional V-ATPase, membrane fusion and endosomal escape of the virus are thought to be precluded [18,28]. It was recently shown that small molecules targeting the V-ATPase improve influenza virus infection outcomes in mice, suggesting that V-type ATPase activity is required for infection in vivo and short-term reduction of V-type ATPase activity in the lungs is tolerated by the host [30]. It was also shown that three host proteins, WDR7, CCDC115, and TMEM199, are required for the assembly of the V-ATPase on the endosome [18]. Loss of these factors promoted over-acidification of incoming endosomes and the lysosomal digestion of endocytosed viral components. These findings have not been tested in vivo, however, and it remains to be determined whether downregulation of these factors would be well-tolerated by the host.
Ubiquitin ligases
Ubiquitin ligases are frequently identified in high-throughput screens with influenza viruses, likely affecting different aspects of the viral replication cycle. One host factor of particular interest is the E3 ubiquitin ligase UBR4. Largescale meta-analysis of RNAi screens with influenza virus paired with interactome data identified UBR4 as an important host factor for replication of human influenza viruses [20]. Mechanistic studies further showed that UBR4 is required for the apical targeting of the influenza A M2 protein. Transient depletion of the UBR4 protein in the lungs prior to infection with influenza A virus improved both morbidity and mortality during infection, though minimal weight loss was observed due to the depletion. Another E3 ubiquitin ligase, ITCH, was also discovered as a host factor for influenza viruses with RNAi-based screening and is required for efficient endosomal escape of influenza virus [12]. However, it remains unclear if ITCH activity is required for infection in vivo. Notably, both ITCH and UBR4 are required for infection with several other clinically relevant RNA viruses [31–33], expanding opportunities beyond influenza viruses for more broad-spectrum antiviral intervention strategies.
Innate immune factors
Perhaps the most well-studied and characterized class of influenza restriction factors are innate immune effector proteins. Both loss-of-function and gain-of-function screening strategies have repeatedly identified the IFITM proteins (specifically IFITM2 and IFITM3), and MxA as strong antagonists of influenza virus infection [9,14,15]. The IFITM proteins are interferon-stimulated genes (ISGs) that generally antagonize viral infections and are canonically thought to inhibit influenza virus entry and endocytosis [34,35]. It was further demonstrated mechanistically that IFITM3 engages binding viral particles and shuttles them to the lysosome to avoid infection [36]. Additionally, multiple groups have shown that endogenously expressed IFITM3 restricts influenza virus infections in vivo [37,38].
The human MX1 gene is an ISG that encodes the MxA protein, a cytoplasmic GTPase. The MxA protein antagonizes influenza virus replication by preventing nuclear import of viral proteins, though the precise mechanism for this is still unclear [39]. Regardless, overexpression of MX1 is sufficient to prevent influenza virus infection in vitro [14]. Importantly, the mouse MX1 gene (encoding the MX1 protein) is distinct from the human orthologue, in that it is localized to the nucleus and inhibits viral transcription [40]. It has been extensively demonstrated that murine MX1, and to a lesser extent human MxA, antagonize influenza virus infection in vivo [39,41,42]. Further, targeted delivery of the murine MX1 protein to mouse lungs using a cell-penetrating peptide strategy protects mice from lethal challenge with influenza A virus [43]. Thus, upregulation of human MX1 or delivery of murine MX1 could be promising avenues to pursue. Finally, the IFITM proteins, human MxA, and murine MX1 have been shown to restrict a wide range of RNA viruses, again demonstrating the opportunity for broad-spectrum antiviral strategies [9,34,35,44].
One overarching question regarding the directed modulation of some innate immune factors, however, is whether the activation of antiviral genes would have a high likelihood of causing undesired side effects. We believe that individual overexpression of ISGs such as MX1, IFITM2, and IFITM3 are more likely to be tolerated compared to the induction of the viral sensors upstream of ISGs. For example, while the RNA-sensor MDA5 helps to restrict influenza virus infection in vivo, its expression is also sufficient for the production of IFN-β, and gain-of-function mutations are associated with autoimmune disorders [15,45]. While transient alterations to gene expression may avoid at least some of these pitfalls, in vivo studies will be required to understand the feasibility of targeting innate immune effectors or regulators either prophylactically or therapeutically.
Strategies for in vivo gene modulation
Even with high-priority host targets defined, mediating effective, safe, and transient alterations to host factor activity is non-trivial. Traditionally, host factors have been downregulated or blocked using small molecule inhibitors and monoclonal antibodies [6]. Despite FDA approval and clinical efficacy in several different contexts, these strategies are met with several challenges. Limitations can include: the broad activity of small molecules delivered systemically, the lack of effective small molecule inhibitors for most genetic targets, and the difficulty of targeting intracellular host-dependency factors with external biologics [46]. Thus, the development of additional gene modulator modalities is likely to facilitate the realization of novel host-directed therapeutics.
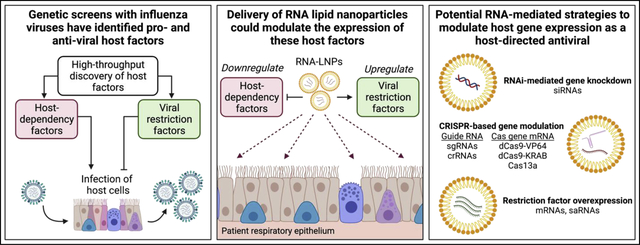
One alternative for up- or down-regulating host factor activities is the direct modulation of host gene expression. Host gene expression can be modulated using a variety of methods spanning from viral vectors to nucleic acid-based technologies [47–51]. One promising approach, the targeted modulation and delivery of host genes in specific tissues using RNAs, has recently developed at an astounding pace. Most notably, the use of mRNA lipid nanoparticles (LNPs) to deliver the SARS-CoV-2 spike gene as a vaccination strategy was proven to be effective and safe at the global scale over the past year [52]. It has further been shown that mRNA-LNPs can efficiently and specifically deliver genes to the tissues of both the upper and lower airways where influenza viruses infect [53,54]. Thus, we will focus the remainder of the review on the potential application of these RNA-based delivery approaches for host-directed antivirals against influenza virus infections, including the various strategies that could be used to overexpress viral restriction factors and downregulate host-dependency factors in the respiratory tract (Figure 1).
Figure 1. Potential approaches for modulating host genes as host-directed antiviral strategies.
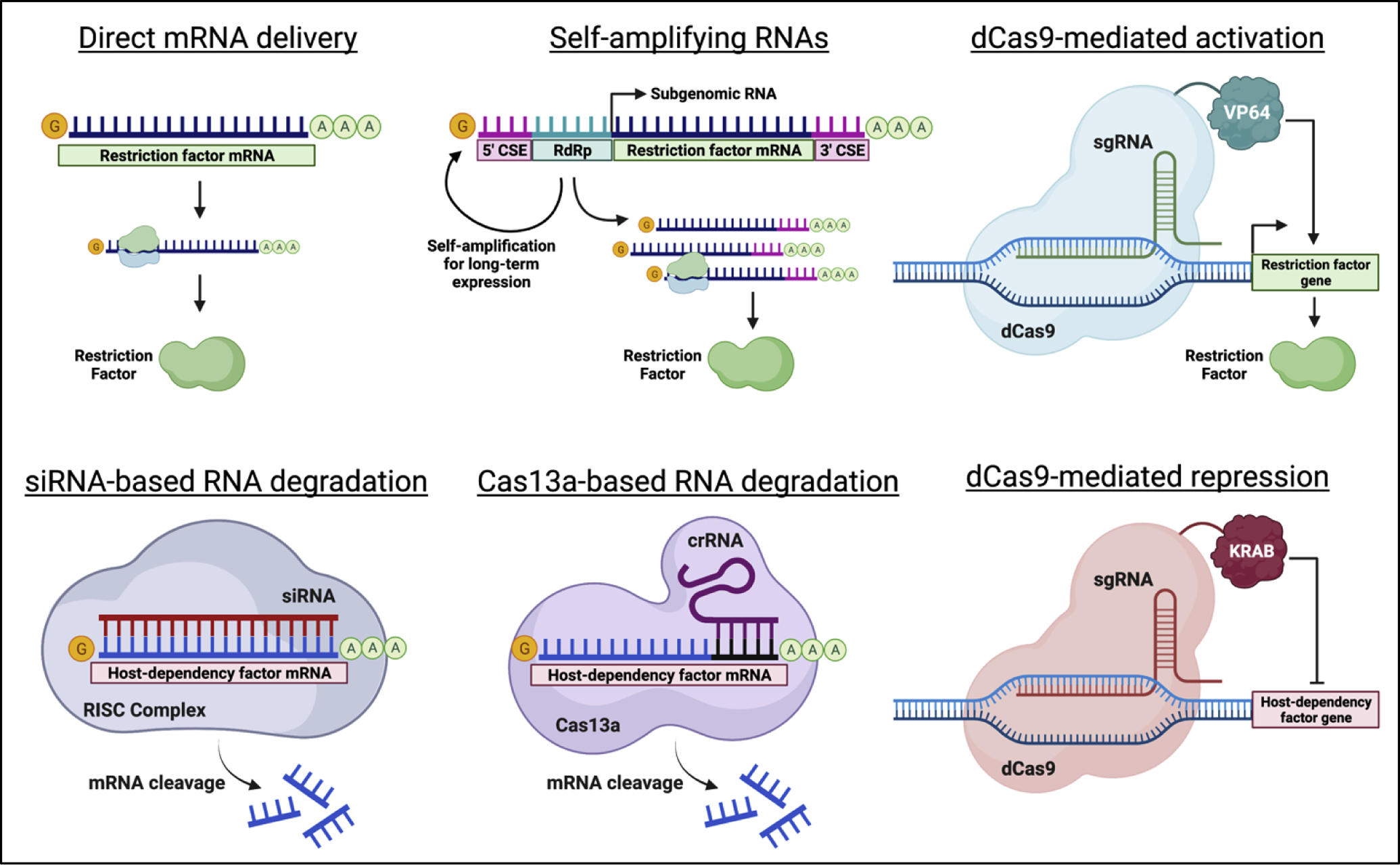
Schematic detailing various RNA-based strategies for the upregulation of host restriction factors (top) or the downregulation of host-dependency factors (bottom) to control viral infections. Abbreviations: crRNA, CRISPR RNA; CSE, alphavirus conserved sequence element; dCas9, nuclease-dead Cas9; RdRp, alphavirus RNA-dependent RNA polymerase; sgRNA, single guide RNA; siRNA, small interfering RNA.
Several approaches for the overexpression of transgenes have been implemented for vaccination strategies and gene therapy. For many applications, delivery of mRNAs encoding the gene of interest is sufficient [55]. Expression of the gene tends to be highly transient, lasting only 24–48 hours with peaks in the first 24 hours after delivery [55,56]. For more long-term approaches, self-amplifying RNAs (saRNAs) and CRISPR-mediated gene activation (CRISPRa) may be more appropriate. saRNAs encode an RNA-dependent RNA polymerase to replicate the RNA delivered to the cell, allowing for high expression of the transgene for up to 60 days [57,58]. CRISPRa can overexpress genes from the host genome directly using nuclease-dead Cas9 protein fused to transcriptional activators such as VP64 [59]. Long-term transcriptional activation of host genes has been achieved through delivery of the required CRISPRa components with adeno-associated viral vectors [60]. More recently, systemic delivery of the CRISPR components with mRNA-LNPs has been achieved for tissue-specific gene editing [61]. It is feasible this technology could be adopted for the expression of CRISPRa machinery in the respiratory tract using similar tactics.
Downregulation of host genes via RNA delivery can also be achieved through a variety of methods. The first demonstration of gene expression modulation in the lungs through RNA was accomplished through siRNA-based technologies [62]. Notably, intranasal delivery of siRNAs targeting the respiratory syncytial virus (RSV) genome conferred protection against RSV in mice and was safely tolerated in human clinical trials [63]. siRNA-based therapeutics may therefore also apply to the downregulation of host factors required for viral replication in the respiratory epithelium. More recently, CRISPR-mediated gene downregulation via CRISPR interference (CRISPRi) and Cas13a-mediated RNA degradation have been described. CRISPRi suppresses gene expression by fusing transcriptional repressors such as the Krüppel-associated box (KRAB) domain to nuclease-dead Cas9 [64]. CRISPRi has been used to suppress gene expression in vivo, though not in the respiratory tract [65]. Cas13a has been successfully used to degrade both influenza and SARS-CoV-2 RNA in the respiratory epithelium in animal models [66]. Similar delivery strategies could therefore be used to target host dependency factors to limit viral replication. Unlike genetic activation strategies, both siRNA-based and Cas13a-mediated degradation of host-dependency factor mRNA could simultaneously target the viral genome directly. In theory, such an approach would further limit viral replication and reduce the probability of viral escape mutants for either antiviral strategy.
Conclusion
Host-directed intervention strategies for influenza virus infections have become increasingly achievable in recent years. While the list of candidate genes presented in this review is far from exhaustive, many of these factors show efficacy in vivo with minimal host toxicity after activity is modulated using various strategies. It is also important to note that while we assessed host factors discovered with high-throughput strategies, several factors have been identified in hypothesis-based studies with equal potential for eventual development of host-directed interventions [67,68]. Future work in the field will focus on modulating the expression of these candidates and others within the respiratory tract, first in animal models and eventually in a clinical setting. Work in this area could contribute not only to influenza virus intervention strategies, but also to the development of truly broad-spectrum antivirals to control communicable respiratory diseases.
Acknowledgements
We apologize to those whose work we could not include due to space constraints. Schematics were created with BioRender.com. NSH is a member of the Duke Cancer Institute and is partially supported by grants from NIAID and NHLBI, grant numbers: R01-HL142985, R01-AI137031. Additionally, N.S.H is funded in part by the Defense Advanced Research Projects Agency’s (DARPA) PReemptive Expression of Protective Alleles and Response Elements (PREPARE) program (Cooperative agreement #HR00111920008). JDT was partially supported by T32-CA009111. The views, opinions and/or findings expressed are those of the authors and should not be interpreted as representing the official views or policies of the U.S. Government.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest
The authors declare no conflicts of interest.
References
- [1].Lafond KE, Porter RM, Whaley MJ, Suizan Z, Ran Z, Aleem MA, Thapa B, Sar B, Proschle VS, Peng Z, et al. : Global burden of influenza-associated lower respiratory tract infections and hospitalizations among adults: A systematic review and meta-analysis. PLoS Med 2021, 18:e1003550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Paget J, Spreeuwenberg P, Charu V, Taylor RJ, Iuliano AD, Bresee J, Simonsen L, Viboud C, Global Seasonal Influenza-associated Mortality Collaborator N, Teams* GLC: Global mortality associated with seasonal influenza epidemics: New burden estimates and predictors from the GLaMOR Project. J Glob Health 2019, 9:020421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Osterholm MT, Kelley NS, Sommer A, Belongia EA: Efficacy and effectiveness of influenza vaccines: a systematic review and meta-analysis. Lancet Infect Dis 2012, 12:36–44. [DOI] [PubMed] [Google Scholar]
- [4].United States Food and Drug Administration: Influenza (Flu) Antiviral Drugs and Related Information. Edited by United States Food and Drug Administration; 2020. [Google Scholar]
- [5].Hussain M, Galvin HD, Haw TY, Nutsford AN, Husain M: Drug resistance in influenza A virus: the epidemiology and management. Infect Drug Resist 2017, 10:121–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6]**.Kumar N, Sharma S, Kumar R, Tripathi BN, Barua S, Ly H, Rouse BT: Host-Directed Antiviral Therapy. Clin Microbiol Rev 2020, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]; Thorough review on host-directed strategies to combat viral infections, with discussion of current FDA-approved therapeutics and approaches still in pre-clinical development.
- [7].Hao L, Sakurai A, Watanabe T, Sorensen E, Nidom CA, Newton MA, Ahlquist P, Kawaoka Y: Drosophila RNAi screen identifies host genes important for influenza virus replication. Nature 2008, 454:890–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Carette JE, Guimaraes CP, Varadarajan M, Park AS, Wuethrich I, Godarova A, Kotecki M, Cochran BH, Spooner E, Ploegh HL, et al. : Haploid genetic screens in human cells identify host factors used by pathogens. Science 2009, 326:1231–1235. [DOI] [PubMed] [Google Scholar]
- [9].Brass AL, Huang IC, Benita Y, John SP, Krishnan MN, Feeley EM, Ryan BJ, Weyer JL, van der Weyden L, Fikrig E, et al. : The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell 2009, 139:1243–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Konig R, Stertz S, Zhou Y, Inoue A, Hoffmann HH, Bhattacharyya S, Alamares JG, Tscherne DM, Ortigoza MB, Liang Y, et al. : Human host factors required for influenza virus replication. Nature 2010, 463:813–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Karlas A, Machuy N, Shin Y, Pleissner KP, Artarini A, Heuer D, Becker D, Khalil H, Ogilvie LA, Hess S, et al. : Genome-wide RNAi screen identifies human host factors crucial for influenza virus replication. Nature 2010, 463:818–822. [DOI] [PubMed] [Google Scholar]
- [12].Su WC, Chen YC, Tseng CH, Hsu PW, Tung KF, Jeng KS, Lai MM: Pooled RNAi screen identifies ubiquitin ligase Itch as crucial for influenza A virus release from the endosome during virus entry. Proc Natl Acad Sci U S A 2013, 110:17516–17521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Tran AT, Rahim MN, Ranadheera C, Kroeker A, Cortens JP, Opanubi KJ, Wilkins JA, Coombs KM: Knockdown of specific host factors protects against influenza virus-induced cell death. Cell Death Dis 2013, 4:e769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Schoggins JW, MacDuff DA, Imanaka N, Gainey MD, Shrestha B, Eitson JL, Mar KB, Richardson RB, Ratushny AV, Litvak V, et al. : Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature 2014, 505:691–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Benitez AA, Panis M, Xue J, Varble A, Shim JV, Frick AL, Lopez CB, Sachs D, tenOever BR: In Vivo RNAi Screening Identifies MDA5 as a Significant Contributor to the Cellular Defense against Influenza A Virus. Cell Rep 2015, 11:1714–1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Heaton BE, Kennedy EM, Dumm RE, Harding AT, Sacco MT, Sachs D, Heaton NS: A CRISPR Activation Screen Identifies a Pan-avian Influenza Virus Inhibitory Host Factor. Cell Rep 2017, 20:1503–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Han J, Perez JT, Chen C, Li Y, Benitez A, Kandasamy M, Lee Y, Andrade J, tenOever B, Manicassamy B: Genome-wide CRISPR/Cas9 Screen Identifies Host Factors Essential for Influenza Virus Replication. Cell Rep 2018, 23:596–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18]*.Li B, Clohisey SM, Chia BS, Wang B, Cui A, Eisenhaure T, Schweitzer LD, Hoover P, Parkinson NJ, Nachshon A, et al. : Genome-wide CRISPR screen identifies host dependency factors for influenza A virus infection. Nat Commun 2020, 11:164. [DOI] [PMC free article] [PubMed] [Google Scholar]; This work provides an extensive meta-analysis of previously completed high-throughput approaches to distill a large amount of data into important concensus influenza virus host-dependency factors, including loss-of-function RNAi and CRISPR knockout screening and proteomics data.
- [19]*.Martin-Sancho L, Tripathi S, Rodriguez-Frandsen A, Pache L, Sanchez-Aparicio M, McGregor MJ, Haas KM, Swaney DL, Nguyen TT, Mamede JI, et al. : Restriction factor compendium for influenza A virus reveals a mechanism for evasion of autophagy. Nat Microbiol 2021, 6:1319–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]; This work constructs a useful influenza virus restriction factor compendium from previously generated loss-of-function high-throughput screening, transcriptomics, and proteomics data.
- [20]**.Tripathi S, Pohl MO, Zhou Y, Rodriguez-Frandsen A, Wang G, Stein DA, Moulton HM, DeJesus P, Che J, Mulder LC, et al. : Meta- and Orthogonal Integration of Influenza “OMICs” Data Defines a Role for UBR4 in Virus Budding. Cell Host Microbe 2015, 18:723–735. [DOI] [PMC free article] [PubMed] [Google Scholar]; This work demonstrates that transient depletion of UBR4, an E3 ubiquitin ligase, in the mouse respiratory epithelium improves survival and morbidity in mice infected with influenza A virus. Additionally, the authors provide a rich resource of validated influenza virus host-dependency and viral restriction factors derived from a meta-omics analysis of previously completed RNAi screens.
- [21].Kumlin U, Olofsson S, Dimock K, Arnberg N: Sialic acid tissue distribution and influenza virus tropism. Influenza Other Respir Viruses 2008, 2:147–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Li Y, Chen X: Sialic acid metabolism and sialyltransferases: natural functions and applications. Appl Microbiol Biotechnol 2012, 94:887–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Schwarzkopf M, Knobeloch KP, Rohde E, Hinderlich S, Wiechens N, Lucka L, Horak I, Reutter W, Horstkorte R: Sialylation is essential for early development in mice. Proc Natl Acad Sci U S A 2002, 99:5267–5270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Jiang J, Sun Y, Xiao R, Wai K, Ahmad MJ, Khan FA, Zhou H, Li Z, Zhang Y, Zhou A, et al. : Porcine antiviral activity is increased by CRISPRa-SAM system. Biosci Rep 2019, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Martins MF, Honorio-Ferreira A, Martins P, Goncalves CA: Presence of sialic acids in bronchioloalveolar cells and identification and quantification of N-acetylneuraminic and N-glycolylneuraminic acids in the lung. Acta Histochem 2019, 121:712–717. [DOI] [PubMed] [Google Scholar]
- [26].Nicholls JM, Moss RB, Haslam SM: The use of sialidase therapy for respiratory viral infections. Antiviral Res 2013, 98:401–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Moss RB, Hansen C, Sanders RL, Hawley S, Li T, Steigbigel RT: A phase II study of DAS181, a novel host directed antiviral for the treatment of influenza infection. J Infect Dis 2012, 206:1844–1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Lakadamyali M, Rust MJ, Zhuang X: Endocytosis of influenza viruses. Microbes Infect 2004, 6:929–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Chou YC, Lai MM, Wu YC, Hsu NC, Jeng KS, Su WC: Variations in genome-wide RNAi screens: lessons from influenza research. J Clin Bioinforma 2015, 5:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Hu CJ, Chen YT, Fang ZS, Chang WS, Chen HW: Antiviral efficacy of nanoparticulate vacuolar ATPase inhibitors against influenza virus infection. Int J Nanomedicine 2018, 13:8579–8593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Baillet N, Krieger S, Carnec X, Mateo M, Journeaux A, Merabet O, Caro V, Tangy F, Vidalain PO, Baize S: E3 Ligase ITCH Interacts with the Z Matrix Protein of Lassa and Mopeia Viruses and Is Required for the Release of Infectious Particles. Viruses 2019, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Han Z, Sagum CA, Bedford MT, Sidhu SS, Sudol M, Harty RN: ITCH E3 Ubiquitin Ligase Interacts with Ebola Virus VP40 To Regulate Budding. J Virol 2016, 90:9163–9171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Morrison J, Laurent-Rolle M, Maestre AM, Rajsbaum R, Pisanelli G, Simon V, Mulder LC, Fernandez-Sesma A, Garcia-Sastre A: Dengue virus co-opts UBR4 to degrade STAT2 and antagonize type I interferon signaling. PLoS Pathog 2013, 9:e1003265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Zhao X, Li J, Winkler CA, An P, Guo JT: IFITM Genes, Variants, and Their Roles in the Control and Pathogenesis of Viral Infections. Front Microbiol 2018, 9:3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Bailey CC, Zhong G, Huang IC, Farzan M: IFITM-Family Proteins: The Cell’s First Line of Antiviral Defense. Annu Rev Virol 2014, 1:261–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Spence JS, He R, Hoffmann HH, Das T, Thinon E, Rice CM, Peng T, Chandran K, Hang HC: IFITM3 directly engages and shuttles incoming virus particles to lysosomes. Nat Chem Biol 2019, 15:259–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Everitt AR, Clare S, Pertel T, John SP, Wash RS, Smith SE, Chin CR, Feeley EM, Sims JS, Adams DJ, et al. : IFITM3 restricts the morbidity and mortality associated with influenza. Nature 2012, 484:519–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Bailey CC, Huang IC, Kam C, Farzan M: Ifitm3 limits the severity of acute influenza in mice. PLoS Pathog 2012, 8:e1002909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Haller O, Kochs G: Mx genes: host determinants controlling influenza virus infection and trans-species transmission. Hum Genet 2020, 139:695–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Pavlovic J, Haller O, Staeheli P: Human and mouse Mx proteins inhibit different steps of the influenza virus multiplication cycle. J Virol 1992, 66:2564–2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Tumpey TM, Szretter KJ, Van Hoeven N, Katz JM, Kochs G, Haller O, Garcia-Sastre A, Staeheli P: The Mx1 gene protects mice against the pandemic 1918 and highly lethal human H5N1 influenza viruses. J Virol 2007, 81:10818–10821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Pavlovic J, Arzet HA, Hefti HP, Frese M, Rost D, Ernst B, Kolb E, Staeheli P, Haller O: Enhanced virus resistance of transgenic mice expressing the human MxA protein. J Virol 1995, 69:4506–4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43]*.Jung HE, Oh JE, Lee HK: Cell-Penetrating Mx1 Enhances Anti-Viral Resistance against Mucosal Influenza Viral Infection. Viruses 2019, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]; This works demonstrates that delivery of the murine MX1 protein to the mouse respiratory tract is well-tolerated by the host and sufficient to improve survival outcomes after influenza A virus infection.
- [44].Verhelst J, Hulpiau P, Saelens X: Mx proteins: antiviral gatekeepers that restrain the uninvited. Microbiol Mol Biol Rev 2013, 77:551–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Dias Junior AG, Sampaio NG, Rehwinkel J: A Balancing Act: MDA5 in Antiviral Immunity and Autoinflammation. Trends Microbiol 2019, 27:75–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Slastnikova TA, Ulasov AV, Rosenkranz AA, Sobolev AS: Targeted Intracellular Delivery of Antibodies: The State of the Art. Front Pharmacol 2018, 9:1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Milone MC, O’Doherty U: Clinical use of lentiviral vectors. Leukemia 2018, 32:1529–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Wold WS, Toth K: Adenovirus vectors for gene therapy, vaccination and cancer gene therapy. Curr Gene Ther 2013, 13:421–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Wang D, Tai PWL, Gao G: Adeno-associated virus vector as a platform for gene therapy delivery. Nat Rev Drug Discov 2019, 18:358–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Nan Y, Zhang YJ: Antisense Phosphorodiamidate Morpholino Oligomers as Novel Antiviral Compounds. Front Microbiol 2018, 9:750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Rajsbaum R: Intranasal Delivery of Peptide-Morpholinos to Knockdown Influenza Host Factors in Mice. Methods Mol Biol 2017, 1565:191–199. [DOI] [PubMed] [Google Scholar]
- [52].Chaudhary N, Weissman D, Whitehead KA: mRNA vaccines for infectious diseases: principles, delivery and clinical translation. Nat Rev Drug Discov 2021, 20:817–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53]**.Lokugamage MP, Vanover D, Beyersdorf J, Hatit MZC, Rotolo L, Echeverri ES, Peck HE, Ni H, Yoon JK, Kim Y, et al. : Optimization of lipid nanoparticles for the delivery of nebulized therapeutic mRNA to the lungs. Nat Biomed Eng 2021, 5:1059–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]; This work demonstrates effective delivery of mRNAs to the mouse respiratory epithelium using lipid nanoparticles. The authors further deliver a secreted hemagglutinin-neutralizing antibody to the respiratory epithelium to protect from a lethal challenge with influenza virus in mice.
- [54].Shaffer C: Mist begins to clear for lung delivery of RNA. Nat Biotechnol 2020, 38:1110–1112. [DOI] [PubMed] [Google Scholar]
- [55].Truong B, Allegri G, Liu XB, Burke KE, Zhu X, Cederbaum SD, Haberle J, Martini PGV, Lipshutz GS: Lipid nanoparticle-targeted mRNA therapy as a treatment for the inherited metabolic liver disorder arginase deficiency. Proc Natl Acad Sci U S A 2019, 116:21150–21159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Pardi N, Tuyishime S, Muramatsu H, Kariko K, Mui BL, Tam YK, Madden TD, Hope MJ, Weissman D: Expression kinetics of nucleoside-modified mRNA delivered in lipid nanoparticles to mice by various routes. J Control Release 2015, 217:345–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Blakney AK, McKay PF, Yus BI, Aldon Y, Shattock RJ: Inside out: optimization of lipid nanoparticle formulations for exterior complexation and in vivo delivery of saRNA. Gene Ther 2019, 26:363–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58]*.Bloom K, van den Berg F, Arbuthnot P: Self-amplifying RNA vaccines for infectious diseases. Gene Ther 2021, 28:117–129. [DOI] [PMC free article] [PubMed] [Google Scholar]; Comprehensive review on the construction of self-amplifying RNA delivery vectors and their use to transiently express antigens for vaccination strategies.
- [59]*.Pandelakis M, Delgado E, Ebrahimkhani MR: CRISPR-Based Synthetic Transcription Factors In Vivo: The Future of Therapeutic Cellular Programming. Cell Syst 2020, 10:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]; Review compiling the various methods for modulating host gene expression using CRISPR-based gene targeting strategies.
- [60].Liao HK, Hatanaka F, Araoka T, Reddy P, Wu MZ, Sui Y, Yamauchi T, Sakurai M, O’Keefe DD, Nunez-Delicado E, et al. : In Vivo Target Gene Activation via CRISPR/Cas9-Mediated Trans-epigenetic Modulation. Cell 2017, 171:1495–1507 e1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Wei T, Cheng Q, Min YL, Olson EN, Siegwart DJ: Systemic nanoparticle delivery of CRISPR-Cas9 ribonucleoproteins for effective tissue specific genome editing. Nat Commun 2020, 11:3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Lam JK, Liang W, Chan HK: Pulmonary delivery of therapeutic siRNA. Adv Drug Deliv Rev 2012, 64:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].DeVincenzo J, Cehelsky JE, Alvarez R, Elbashir S, Harborth J, Toudjarska I, Nechev L, Murugaiah V, Van Vliet A, Vaishnaw AK, et al. : Evaluation of the safety, tolerability and pharmacokinetics of ALN-RSV01, a novel RNAi antiviral therapeutic directed against respiratory syncytial virus (RSV). Antiviral Res 2008, 77:225–231. [DOI] [PubMed] [Google Scholar]
- [64].Alerasool N, Segal D, Lee H, Taipale M: An efficient KRAB domain for CRISPRi applications in human cells. Nat Methods 2020, 17:1093–1096. [DOI] [PubMed] [Google Scholar]
- [65].MacLeod RS, Cawley KM, Gubrij I, Nookaew I, Onal M, O’Brien CA: Effective CRISPR interference of an endogenous gene via a single transgene in mice. Sci Rep 2019, 9:17312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66]*.Blanchard EL, Vanover D, Bawage SS, Tiwari PM, Rotolo L, Beyersdorf J, Peck HE, Bruno NC, Hincapie R, Michel F, et al. : Treatment of influenza and SARS-CoV-2 infections via mRNA-encoded Cas13a in rodents. Nat Biotechnol 2021, 39:717–726. [DOI] [PubMed] [Google Scholar]; In this work, authors effectively deliver nebulized mRNA lipid nanoparticles encoding Cas13a to the respiratory epithelium to specifically degrade influenza and SARS-CoV-2 genomic RNA in mice and hamsters, respectively.
- [67].Ehre C, Worthington EN, Liesman RM, Grubb BR, Barbier D, O’Neal WK, Sallenave JM, Pickles RJ, Boucher RC: Overexpressing mouse model demonstrates the protective role of Muc5ac in the lungs. Proc Natl Acad Sci U S A 2012, 109:16528–16533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Halstead ES, Umstead TM, Davies ML, Kawasawa YI, Silveyra P, Howyrlak J, Yang L, Guo W, Hu S, Hewage EK, et al. : GM-CSF overexpression after influenza a virus infection prevents mortality and moderates M1-like airway monocyte/macrophage polarization. Respir Res 2018, 19:3. [DOI] [PMC free article] [PubMed] [Google Scholar]


