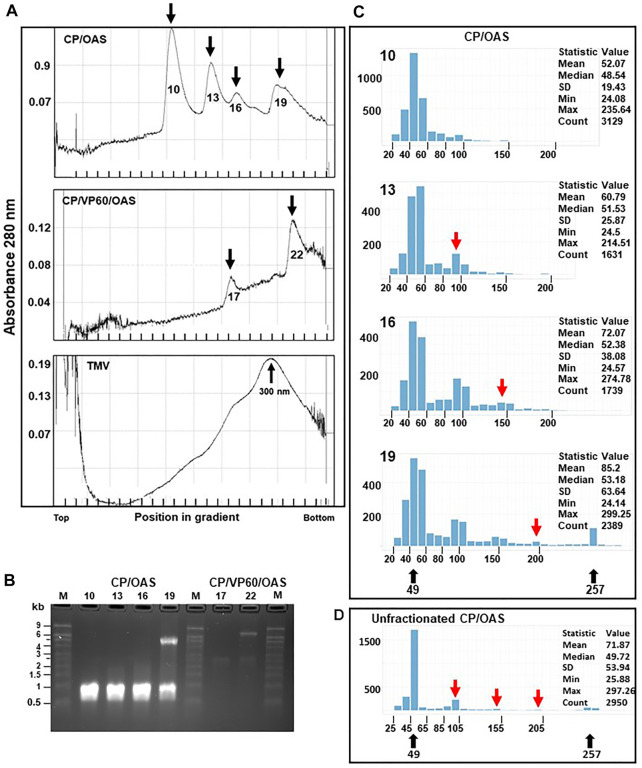
FIGURE 3.
Distinctive size classes of nanorods can be separated by sucrose-density gradient ultracentrifugation centrifugation. (A) CP/OAS nanorods, upper panel, CP/VP60/OAS nanorods, middle panel and TMV lower panel separated on a 15–30% sucrose density gradient and fractionated. Y axes, absorbance (280 nm) measured during fractionation. Fractions, 10, 13, 16 and 19 (X axis) corresponding to measurement peaks are marked by arrows, upper panel, and similarly fractions 17 and 22, middle panel. Lower panel, TMV with the virus peak migration indicated at 300 nm. (B) Ethidium bromide-stained denaturing agarose gel of RNA extracted from fractions indicated in (A). M, RNA size ladder. (C) Histograms, output of the Nanorod UI interface, showing the length distribution of nanorods from computer-automated measurements of TEM images for CP/OAS fractions 10, 13, 16 and 19. The nanorod expected lengths 49 and 257 nm are indicated below. Y axes, number of nanorods in each size class; X axes, length distributions in 10 nm increments. The expected length of nanorods derived by end-to-end formation of 49 nm nanorods is shown, red arrows. The descriptive measurement statistics (including mean, median, standard deviation; minimum and maximum length and total count) is shown for each histogram. (D) Histogram showing the length distribution of particle lengths from computer-automated measurements of TEM images for CP/OAS unfractionated nanorods.