FIGURE 4.
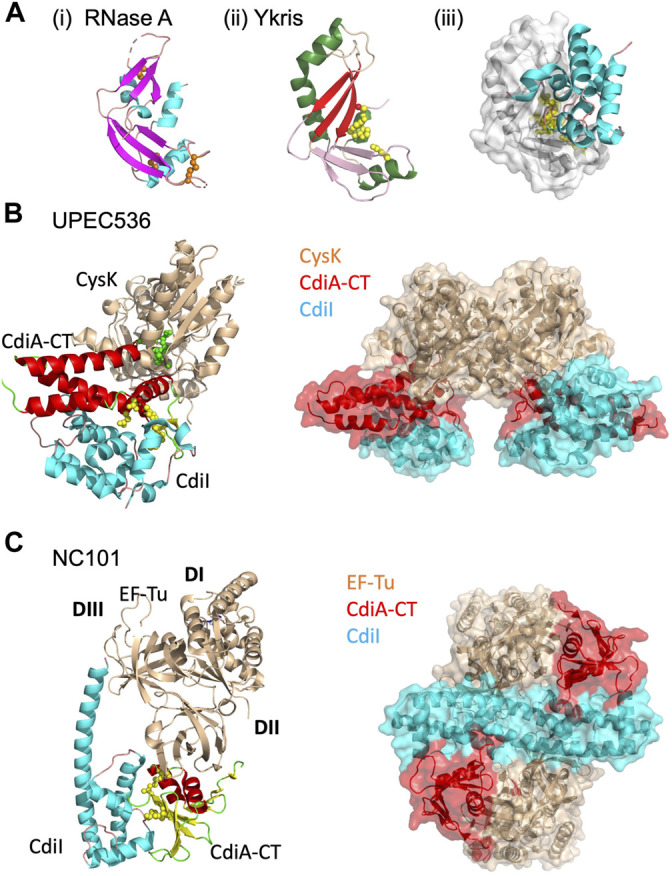
CdiA-CT toxins with unique characteristics. (A) Y. kristensenii (Ykris) [PDB ID: 5E3E (Batot et al., 2017)] has some structural homology to BECR family members, but most closely resembles RNase A [PDB ID: 4B36 (Thiyagarajan and Acharya, 2013)]. i) RNase A is shown in cartoon depiction with β-strands in magenta, α-helices in cyan, and loops in salmon, and with conserved disulfide bonds shown as orange spheres. ii) CdiA-CTYkris in cartoon representation with its BECR core structure colored by as in Figure 2 left panels. iii) The CdiA-CT/CdiIYkris complex with CdiI colored as in Figure 2 right panels. In (B,C) the accessory protein (CysK or EF-Tu) is shown in beige, while on the left the toxin and immunity protein are colored by secondary structure where CdiA-CT has β-strands in yellow, α-helices in red, and loops in green; CdiI has α-helices in cyan and loops in pink. Active site residues for CdiA-CT are shown as yellow spheres. In the complex on the right, CdiA-CT is colored red, CdiI cyan and the accessory protein is in beige. (B) In UPEC 536, CdiA-CT forms a complex with CdiI and CysK [PDB ID: 5J5V (Johnson et al., 2016a)]. The C-terminal residue of CdiA-CT (green spheres) that inserts into the CysK active site is highlighted. Shown on the right is the overall oligomerization of the CdiA-CT/CdiI/CysK complex, where CysK dimerization results in a dimer of heterotrimers. (C) In E. coli NC101, CdiA-CT interacts directly with CdiI and domain 2 (DII) of EF-Tu, while CdiI interacts with DII and domain 3 (DIII) of EF-Tu (PDB ID: 5I4R; (Jones et al., 2017). On the right, the CdiA-CT/CdiI/EF-Tu complex is shown, where CdiI forms the center of the dimer of heterotrimers.
