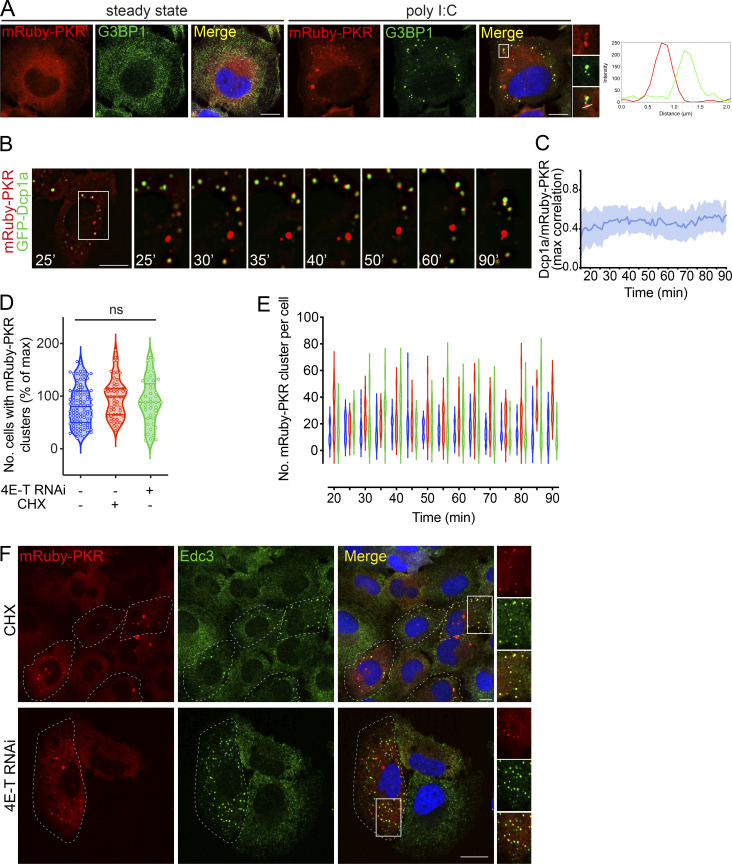
Figure 2.
PKR clusters are autonomous and recruit PB components. (A) Representative immunofluorescence images showing that mRuby-PKR clusters and G3BP1, an SG component, do not colocalize. Right: Plot of signal intensity of mRuby-PKR clusters (red) or G3BP1 immunostaining (green) as a function of distance. The ROI used for metrics is indicated with a white line on the micrograph crops. Scale bars: 10 µm. (B) Time-lapse micrographs showing colocalization and subsequent demixing of mRuby-PKR and GFP-Dcp1a, a PB component. Scale bar: 10 µm. (C) Quantification of the data in B (mean and 95% confidence interval bands; N = 3 experiments, n = 30 cells). (D) Violin plots showing the total number of cells with mRuby-PKR clusters after administration of poly I:C (blue), poly I:C and CHX (red), and poly I:C and 4E-T RNAi (green); N = 3 experiments, n > 2,000; one-way ANOVA. (E) Violin plots showing the number of mRuby-PKR clusters per cell over time in cells treated with poly I:C (blue), poly I:C and CHX (red), and poly I:C and 4E-T RNAi (green); N = 3 experiments, n > 200. (F) Representative micrographs showing that mRuby-PKR clusters recruit the PB component Edc3 after depletion of PBs with CHX and 4E-T RNAi. Scale bar: 10 µm.