Abstract
The glutathione-S-transferase (GST) super family comprises multiple isozymes (Alpha, Mu, Pi, Omega, Theta, and Zeta) with compelling evidence of functional polymorphic variation. Over the last two decades, a significant body of data has accumulated linking aberrant expression of GST isozymes with the development and expression of resistance to cancer drugs. Clinical correlation studies show that genetic differences within the human GST isozymes may play a role in cancer susceptibility and treatment.
The initial confusion was presented by the fact that not all drugs used to select for resistance were substrates for thioether bond catalysis by GSTs. However, recent evidence that certain GST isozymes possess the capacity to regulate mitogen activated protein kinases presents an alternative explanation. This dual functionality has contributed to the recent efforts to target GSTs with novel small molecule therapeutics.
While the ultimate success of these attempts remains to be shown, at least one drug is in late-stage clinical testing. In addition, the concept of designing new drugs that might interfere with protein:protein interactions between GSTs and regulatory kinases provides a novel approach to identify new targets in the search for cancer therapeutics.
1. Glutathione-S-Transferase (GSTs) and Anticancer Drug Response
In cancer chemotherapy, development of drug resistance is a key element in the eventual failure of effective therapeutic treatments. In both preclinical models and in patients, exposure to anticancer agents can provide the selective pressure, which leads to induced expression of protective gene products. Although the drug-resistant phenotype is frequently characterized by multiple and pleiotropic changes, one frequent adaptation is altered expression of glutathione-S-transferases (GSTs).[1,2] GSTs are a family of phase II detoxification enzymes that have as a primary function the protection of cellular macromolecules through catalyzing the conjugation of glutathione (GSH) to a wide variety of endogenous and exogenous electrophilic compounds. While GSTs play a quintessential role in protecting cells from environmental and oxidative stress, they can provide an obstacle to the successful treatment of patients with cancer. For example, a survey of the National Cancer Institute cancer drug screening panel of cell lines showed an inverse correlation between GST expression and sensitivity toward alkylating agents.[3]
Table I provides a list of anti-cancer agents for which resistance has been associated with elevated levels of GSTs. Some of these drugs are substrates of GSTs and can be directly inactivated through catalytic thioether bond conjugation to GSH. However, an early conundrum provided by the GST literature was that many tumor cells over-expressed GST isozymes even if resistance was not to a drug that could act as a substrate. Such apparently conflicting information has been explained by recent studies that have identified new functions for old enzymes. This review will summarize those issues most pertinent to understanding the genetic differences in human GST expression and their relevance to cancer susceptibility and treatment.
Table I.
Anti-cancer agents associated with increased levels of glutathione-S-transferase (GST) and resistance (adapted from Tewi[2] and Fan and Chambers[4])
| Agent | |
|---|---|
| Substrates of GST and thereby inactivated via GSH-conjugation | Chlorambucil |
| Acrolein | |
| Hydroxyalkenals | |
| Carmustine | |
| Nitrogen mustard | |
| Melphalan | |
| Ethacrynic acid | |
| Corticosteroids | |
| Phosphoramide mustard | |
| Non-substrates | Bleomycin |
| Hepsulfam | |
| Carboplatin | |
| Non-substrates but require JNK activation to elicit cytotoxicity | Antimetabolites |
| Antimicrotubule drugs | |
| Topoisomerase I and II inhibitors | |
| Mitomycin C | |
| Adriamycin | |
| Cisplatin |
GSH = glutathione; JNK = c-Jun N-terminal kinase.
2. GSTs and Kinase Regulation
Arguably, one of the more interesting developments in understanding the role of phase II metabolism enzymes in maintaining cellular homeostasis is the recent publication of evidence linking GSTs with kinase-mediated signaling cascades. At this stage, it is premature to offer conjecture on the generality of this regulatory pathway, however, at least two examples of non-catalytic functions for GST isozymes now exist.
Mechanistically, GSTs play a regulatory role in kinase signaling by forming ligand-binding interactions with critical cellular kinases involved in the regulation of proliferation and apoptosis. Through protein : protein interactions, GSTs function to sequester signaling kinases and act as negative regulators. It is ironic to reflect that the recently reported protein ligand binding of the GSTP1–1 isozyme with c-Jun N-terminal kinase (JNK) and of the GSTM1 isozyme with apoptosis signal-regulating kinase (ASK1) present properties reminiscent of the ‘ligandin’ functionality ascribed to the capacity of liver GST to bind reversibly to heme and bilirubin.[5] Thus far, two distinct interactions have been characterized. For example, GSTM1 plays a regulatory role in the heat shock-sensing pathway by binding to, and inhibiting, the activity of ASK1.[6,7] ASK1 is a mitogen-activated protein (MAP) kinase that activates the JNK and p38 pathways leading to cytokine- and stress-induced apoptosis.[8] The activity of ASK1 is low in non-stressed cells due to its sequestration via protein:protein interactions with GSTM1 to form a GSTM1:ASK1 complex. Oxidative stress and heat shock lead to the dissociation of the GSTM1:ASK1 complex resulting in liberation and activation of ASK1.[7,9] Forced expression of GSTM1 blocked ASK1 oligomerization and repressed ASK1-dependent apoptotic cell death.[6] GSTM1 expression is altered in a variety of tumor types and is associated with impaired clinical response to therapy. Thus, in addition to any role that GSTM1 may play in catalyzing GSH conjugation to anti-cancer agents (and this is likely to be minor), it may also influence the apoptotic response through kinase regulation.
The other published example of kinase regulation is provided by the GSTP1 family that plays an integral role in controlling stress response, apoptosis and cellular proliferation through interacting with JNK. JNK has been implicated in pro-apoptotic signaling and may be required for the induced cytotoxicity of a variety of chemotherapy agents, including several of those listed in table I.[4,10] JNK activity is propagated through phosphorylation of c-jun and subsequent downstream effectors. In non-stressed cells, low JNK1 catalytic activity is maintained as a consequence of the sequestration of the protein in a GSTP1:JNK complex.[11,12] However, under conditions of oxidative stress (e.g. ultraviolet [UV] irradiation or hydrogen peroxide treatment) a dissociation of the GSTP1:JNK complex occurs, producing oligomerization of GSTP1 and subsequent induction of apoptosis.[11]
Additional support for this model of GST regulation is provided by the observations that either immunodepletion of GSTP1, or its inhibition by a rationally designed GSH-based peptidomimetic inhibitor, γ-glutamyl-S-(benzyl) cysteinyl-R(−)-phenyl glycine diethyl ester (TLK199), also results in the activation of JNK.[11,12] Collectively, these data provide a plausible explanation of why GST over-expression is a mechanism of drug resistance when the selecting drug is not a substrate for GSH conjugation.
The rational design and synthesis of TLK199 was based on the principle that modulation of drug resistance could be utilized as a viable clinical approach to cancer treatment. While the protein-protein interactions of GSTP1 and JNK have been shown to have a binding constant of approximately 200 nmol/L, the exact site(s) of interaction has not been characterized, other than to implicate the C-terminal end of JNK.[13] It is possible that polymorphisms within each isozyme class may produce distinct binding constants and thereby alter kinase signaling kinetics. It is also apparent that the GSTM1 null phenotype would imply that the mechanism for negative regulation is not universal. However, it is possible for other members of the GST family to substitute for Mu deficiency, illustrating, once again, the potential importance of functional redundancy in GSTs. By implication, the non-enzymatic roles for GSTs in regulating drug response might be a significant consideration when analyzing what impact GST polymorphisms may have in determining cellular response to drug exposure.
3. The GST Gene Super Family: Classification
Human GSTs are divided into two distinct super family members: membrane bound microsomal and cytosolic. Microsomal GSTs contain three isoforms designated mGST 1, 2, and 3 that are encoded by a single gene located on chromosome 12 (MGST1).[14,15] Like cytosolic GSTs, the microsomal isozymes catalyze the conjugation of GSH to electrophilic compounds. Microsomal GSTs play a key role in the endogenous metabolism of leukotrienes and prostaglandins.[14]
The cytosolic GSTs are subject to significant genetic polymorphism in human populations. They are divided into 6 classes that share ~30% sequence identity: Alpha, Mu, Omega, Pi, Theta, and Zeta (see table II). Multiple alleles exist within each class and these share >50% sequence identity (table III).[16] The 5′ promoter region varies between classes and can contain one or more of the following response elements: the antioxidant-response element, the xenobiotic response element, the GSTP enhancer 1, the glucocorticoid-response element, and the Barbie box element.[17–19] Also, the promoter region contains putative binding sites for transcription factors including, AP-1, MAF, Nrfl, Jun, Fos, and NF-kappa B.[18] It is important to note that the occurrence and/or prevalence of these elements are quite species specific and there are particular differences between rodents and humans. As such, while these features provide an adaptive response mechanism to up-regulate GST expression following cellular stress and exposure to toxic xenobiotics, the data do not always lend themselves to extrapolation from mouse to man.
Table II.
Cytosolic glutathione-S-transferases (GSTs)
| Class | Gene(s) | Protein (MW) | Chromosome location |
|---|---|---|---|
| Alpha (α) | GSTA1–5 | 25 900 | 6 |
| Mu (μ) | GSTM1–5 | 26 000–26 700 | 1 |
| Omega (ω) | GSTO1–5 | 27 566 | 10 |
| Pi (π) | GSTA1 | 24 700 | 11 |
| Theta (θ) | GSTA1–2 | 25 100 | 22 |
| Zeta (ζ) | GSTZ1 | 25 000 | 14 |
Table III.
Genetic variation in selected glutathione-S-transferases (GSTs)
| Gene | Allelea | Amino acid change | Enzyme activity | References |
|---|---|---|---|---|
| GSTA1 | *A | Reference | Reference | 27 |
| *B | Pt mutation, promotor | Decreased expression | 27 | |
| GSTA2 | *A | Thr112;Glu210 | Reference | 35 |
| *B | Ser112;Ala210 | No change | 36 | |
| GSTM1 | *A | Lys173 | Reference | 49 |
| *B | Asp173 | No change | 49 | |
| *O | Deletion | None | 50 | |
| *Ax2 | Duplication | Hyper | 51 | |
| GSTM3 | *A | Reference | Reference | 63,64 |
| *B | 3 bp deletion | Altered transcription factor recognition | 65 | |
| GSTM4 | *A | Tyr2517 | Reference | 72 |
| *B | Cyt2517 | No change | 72 | |
| GSTO1 | *A | Ala140;Glu155 | Reference | 80 |
| *B | Ala140;155 deleted | Increased | 80 | |
| *C | Asp140;Glu155 | No change | 80 | |
| GSTP1 | *A | Ile105;Ala114 | Reference | 87,89 |
| *B | Val105;Ala114 | Decreased | 87,89 | |
| *C | VaM05;VaM14 | Decreased | 87,89 | |
| *D | Ile105;Val114 | No change | 88,89 | |
| GSTT1 | *A | Thr104 | Reference | 98,101 |
| *B | Pro104 | Deceased | 98,101 | |
| *O | Deletion | None | 98 | |
| GSTT2 | *A | Met139 | Reference | 98 |
| *B | Ile139 | Not characterized | 98 | |
| GSTZ1 | *A | Lys32;Arg42;Thr82 | Reference | 107,108 |
| *B | Lys32;Gly42;Thr82 | Decreased | 108,109 | |
| *C | Glu32;Gly42;Thr82 | Decreased | 108,109 | |
| *D | Glu32;Gly42;Met82 | Decreased | 107 |
Enzyme activity is measured within each gene using *A, the first allele identified, as a reference.
Cytosolic GSTs function as homo- and hetero-dimeric proteins, allowing the formation of a larger number of enzymes from a limited number of genes, however, dimerization is limited to subunits within the same class.[20] The subunits range in size from 24 to 29 kDa.[21] Each subunit contains an active site with two subsites: a highly conserved G site for GSH binding and an H site for hydrophobic substrates.
Less than 10% of the protein is strictly conserved, and yet all GST isozymes have two domains and a similar topology. The N-terminal domain (residues 1–80) comprises one-third of the protein and forms the G site. It is composed of four β sheets with three flanking α helices, a structural motif common to thioredoxin and other proteins evolved to bind GSH or cysteine.[22] This region contains a catalytically essential tyrosine, serine or cysteine residue that interacts directly with the thiol group of GSH.[22] The C-terminal domain (residues 87–210) is α helical and together with a loop from the N-terminal domain forms the H site. Amino acid variation in the H site accounts for substrate specificity. In addition, structural variations within the C-terminus exist in the Alpha, Theta, and Mu classes. Specifically, an additional C-terminal α helix is present in the Alpha and Theta classes while the Mu class has an extra loop.[22] Both differences are located proximal to the H site, creating a more constricted active site.
Although GSTs are ubiquitously expressed their tissue distribution in mammals is complex. Fetal tissues contain a GST expression profile that is distinct from adults and within some organs, such as the kidney, there are different isoforms expressed even between cell types.[23,24] Adding to the complexity, GSTs have been shown in rodent models to be induced by structurally unrelated compounds known to result in chemical stress and carcinogenesis including: phenobarbital, planar aromatic compounds, ethoxyquin, butylated hydroxyanisol (BHA), and trans-stilbene oxide.[25] Some of the compounds known to induce GSTs are themselves substrates for the enzyme, suggesting that induction may be an adaptive response mechanism.
4. Genomic Considerations: Human Polymorphisms
4.1. Alpha Class
The Alpha class of GSTs is the major isoform expressed in the liver. In fact, an increase in GSTA1*A concentration in blood is a specific marker of hepatocellular impairment.[26] Five genes have been identified in a cluster on chromosome 6 that encode proteins belonging to the Alpha class (GSTA1, A2, A3, A4 and A5).[27–30] GSTA1, GSTA2 and GSTA4 are widely expressed in human tissues, whereas GSTA3 is rare and GSTA5 was not detected in any tissues examined.[27] The GSTA1 and GSTA2 genes span a region of ~12kb and contain 7 exons.[31,32] GSTA1 contains 8 single nucleotide polymorphisms. A silent mutation in exon 5 (A375G) and seven other single nucleotide changes in the promoter region, none of which appear to alter function, have been identified.[27,33] However, the promoter region of GSTA1*A and GSTA1*B differ by 3 base substitutions at positions −567, −69, and −53 which alter expression.[27] Specifically, a G→A change at position −52 alters binding of Sp1, rendering the GSTA1*A promotor more active and therefore more highly expressed.[27] GSTA1 was originally described as ligandin based on its ability to bind to a number of electrophilic compounds.[5] Polymorphisms within this class that lead to differences in expression may alter an individual’s capacity to metabolize drugs and xenobiotics as well as to sequester molecules that may alter kinase signaling. Supporting these conclusions, GSTA1*B gene shows a decreased hepatic expression and appears to confer susceptibility to colon cancer.[34]
The gene encoding GSTA2 was cloned in 1987, and the protein was shown to contain 221 amino acids with a molecular mass of 25 kDa.[28] GSTA2 has two polymorphisms: Thr112/Glu210 and Ser112/Ala210.[35,36] The structure and function of these two isozymes appear identical.[37] As part of the conditioning process, patients undergoing hematopoietic stem cell or bone marrow transplantation are pretreated with the myelosuppressive drug busulfan. While busulfan conjugation to GSH occurs primarily through GSTA2, polymorphisms at this locus appear to have no impact on its biotransformation.[33]
GSTA3 and GSTA4 were identified using the Expressed Sequence Tag database and shown to share approximately 93% and 52% nucleotide sequence identity with GSTA1, respectively.[38] Full-length GSTA3*C was recently cloned and shown to catalyze the double bond isomerization in the biosynthetic pathway of steroid hormones.[39] The isomerase activity in GSTA3 is distinct from other GSTs. Board[38] reported that GSTA3 was a rare transcript. More recent studies show that GSTA3*C is expressed only in tissues characterized by active steroid hormone biosynthesis, including testis, ovary, adrenal gland, and placenta.[39] Polymorphism within GSTA3 and the contribution this subclass may have in the pathology of hormone producing tissues has not been investigated.
GSTA4 was isolated and cloned from a human adult brain cDNA library and shown to have a high activity with reactive carbonyl compounds, including alkenals.[38] The expression has been examined in normal and pathological human tissues and shown to be widely expressed throughout many organs, including the liver, kidney, colon, heart, brain, and skin.[19] In rat neuronal tissue, GSTA4 activity increases with age; however these studies have not been extended to the human isoform.[40] Increased expression of GSTA4 was observed in tissues damaged from reactive oxygen species, including the liver, UV-irradiated skin and the heart. However, expression was decreased in hepatocellular carcinoma.[19] In a separate study using a mouse model, these authors showed that GSTA4 expression is induced in the liver and kidney following iron overload.[41] Based on these two studies, these authors conclude that GSTA4 expression may increase with the formation of free radicals.
The role of GSTA4 and GSTA5 in response to chemotherapeutic agents has been investigated with mouse and rat homologs.[42,43] GSTA4 has been shown to confer resistance to doxorubicin in Chinese hamster ovary cells.[43] The mechanism of resistance was attributed to the inactivation of lipid peroxidation products via GSTA4. Resistance to doxorubicin and other alkylating agents was observed in a hamster fibroblast cell line transfected with the GSTA5 gene.[42] Induction of GSTA5 in rat hepatocytes was observed following exposure of chemotherapy agents.[44] Computer modeling of the promotor region in rat GSTA5 identified a putative antioxidant response element that may be responsible for the induction of this isozyme by chemotherapeutic agents.[44,45] The information on GSTA4 and A5 is limited in human studies, however, rodent models suggest that GSTA may play a role in clinical response to therapeutic agents.
4.2. Mu Class
Five genes have been identified that encode proteins belonging to the Mu class (GSTM1-5).[46] GSTM proteins are encoded by a gene cluster located on chromosome 1.[46,47] Four of the GSTM genes are spaced ~20kb apart in the following orientation; 5′ GSTM4—GSTM2—GSTM1—GSTM5 3′.[48] The GSTM1 gene contains four alleles and has been the most widely studied member of the class (table IV). GSTM1*A and *B differ by one amino acid change (table III) and are enzymatically identical.[49] GSTM1*A has been associated with a decreased risk of bladder cancer and has an allele frequency of 20%.[50] Some Saudi Arabian individuals have demonstrated an enhanced GSTM1 enzyme activity that has been characterized as a gene duplication at this locus.[51] The frequency of this genotype and corresponding risk assessment has not yet been defined.
Table IV.
| Allele | Frequency in Caucasian populations | Frequency in colorectal cancer patients | Cancer risks |
|---|---|---|---|
| M1*A | 0.2 | 0.19 | Decreased risk of bladder and breast cancer |
| M1*B | 0.2 | 0.14 | Decreased risk of pituitary adenomas |
| M1*O | 0.59 | 0.65 | Increased risk of lung, colon, bladder, and post-menopausal breast cancer |
GSTM1 activity is absent in a large number of individuals due to a gene deletion. GSTM1 and M2 genes are in close physical proximity and share 99% nucleotide sequence identity. It is proposed that an unequal crossing over of these two genes resulted in a 15kb gene deletion (the GSTM1*0 allele).[46] GSTM1*0 is surprisingly common, with an average frequency of 50% in human populations with a range of 22% in Nigerians to 67% in Australians.[50]
The GSTM1 null phenotype has been extensively studied as a risk factor for a variety of cancers as these individuals may be subject to an increased sensitivity to carcinogens.[52,53] The null phenotype (homozygous GSTM1*0) is associated with an increased risk of lung, colon, head and neck, and bladder cancer, and aplastic anemia, and is a risk factor for pulmonary asbestosis.[54,55]
The literature defining the role that GSTM1*0 plays in response to chemotherapy agents is contradictory. Patients with breast cancer (homozygous GSTM1*0) treated with cyclophosphamide and adriamycin had a reduced risk of recurrence compared with patients with a wild-type phenotype.[56] Yet, patients with ovarian cancer with a null phenotype treated with alkylating agents showed a poorer prognosis.[57] Defining a causal relationship between the GSTM1*0 phenotype and risk assessment in colorectal cancer is also contradictory.[58,59] However, more recent studies show that individual susceptibility to colorectal cancer was increased when patients expressed both GSTM3*B and GSTM1*0.[60] These data support the general conclusion that the individual phenotype for the GST family members as a whole, rather than a single isoform, should be considered when viewing risks and outcome.
Neurodegenerative diseases such as Parkinson’s disease and schizophrenia are characterized by the degeneration of dopaminergic neurons. Cytosolic prostaglandin E synthase was identified in human brain and characterized as GSTM2.[61] GSTM2*B has been shown to catalyze the conjugation of GSH to aminochrome, a reactive oxygen species generated in the redox cycling of orthoquinones within dopaminergic neurons.[62] Hence, GSTM2*B has been proposed to play a protective role against neurodegenerative diseases.
GSTM3 was isolated in brain extracts and later shown to be expressed in brain and testis.[63,64] The GSTM3 locus contains 2 alleles, *A and *B. The GSTM3*B allele has a three base pair deletion in intron 6 that introduces a recognition sequence for the multifunctional transcription factor YY1.[65] YY1 has been shown to activate and repress transcription thereby altering many cellular responses.[66] The GSTM3*AA genotype was shown to occur more frequently in patients with multiple cutaneous basal cell carcinoma than GSTM3*BB.[67] GSTM3*AA is also associated with an increased risk for laryngeal squamous cell carcinoma, while GSTM3*BB was putatively protective.[68] In contrast, GSTM3*A and *B are expressed in brain, however, there appears to be no direct relationship between GSTM3 and the incidence of astrocytomas.[69]
GSTM4 was cloned and shown to be 87% identical with GSTM1.[70,71] The GSTM4 locus also contains two alleles, *A and *B.[72] The GSTM4*B allele has been recently implicated as a risk factor in the development of lung cancer.[73] The Mu class of GSTs was analyzed in leukemic blasts from 21 children with acute lymphoblastic leukaemia (ALL).[73] GSTM3 and GSTM4 were expressed in 62% and 24% of patients, respectively.[73] These studies showed that GSTM3 was positively related to good prognosis and further information on this class might provide more information for treatment of children with ALL.
4.3. Omega Class
The Omega class of GSTs has recently been described and contains two members, GSTO1 and GSTO2. While this class shares sequence similarity with other GSTs, it is important to recognize that they are structurally and functionally distinct. The GSTO1*A protein is encoded by a single gene on chromosome 10 and is expressed abundantly in liver, macrophages, glial and endocrine cell.[74] GSTO1*A has been shown to be up-regulated in estrogen receptor-negative human breast cancer cell lines.[75] GSTO1*A has 2 unique features that may define this family, separating it from other eukaryotic GSTs. First, using X-ray crystallography, a 19 residue N-terminal extension has been identified that forms a novel structural unit, the function of which remains unclear.[74] Secondly, known substrates of other GSTs are not turned over by GSTO1*A. However, GSTO1*A demonstrated a GSH-dependent reduction of dehydroascorbate, a function characteristic of glutaredoxins rather than GSTs.[74,76] These issues have made the identity and characterization of GSTO1*A an anomaly. Adding to the complexity, a protein previously described as nuclear chloride channel, NCC27, was shown to share sequence homology to the GSTO1 family.[77] Like GSTO1*A, NCC27 has the N-terminal extension and lacks typical transferase activity. NCC27 is co-localized in cardiac and skeletal muscles with ryanodine receptors (RyRs), which are calcium-releasing channels. The addition of GSTO1*A to a cytoplasmic solution inhibited the activity of RyR2 by 50%.[77] These data along with the ubiquitous expression suggests that GSTO1 may have a fundamental, yet uncharacterized, role in cellular calcium homeostasis.
GSTO1*A was originally identified as the human monomethylarsonic acid reductase, (MMA[V]), and described as being the rate-limiting enzyme of inorganic arsenic metabolism.[78] More recently, variations within GSTO1 have been identified that might contribute to an individual’s ability to metabolize arsenic.[79] Specifically, thioltransferase activity is decreased 75% in the Ala140Asp, and 40% in the Thr217Asn GSTO1 variant, compared with the wild-type GSTO1*A.[79]
Three alleles have been identified in the GSTO1 class; GSTO1*A, GSTO1*B and GSTO1*C.[80] Among the Australian, African, and Chinese populations, GSTO1*A was the most prevalent haplotype with a frequency ranging from 0.6–0.9; while GSTO1*B*A was the least common, with a frequency ranging from 0.01–0.05.[80] The impact of heterogeneity within this class has yet to be defined. However, we do know that the thioltransferase and GSH-conjugation activity among the haplotypes are equivalent for GSTO1*A and *C, however, GSTO1*B had significantly higher activity despite the deletion of E155 (Glu155).[80]
GSTO2 has been recently identified and been shown to share 64% amino acid identity with GSTO1.[80] GSTO2 is separated from GSTO1 by 7.5kb on chromosome 10.[80] GSTO2 expression was most abundant in testis, however, it was observed in a variety of tissues including liver, kidney, skeletal muscle, and prostate.[80] The function of GSTO2 has not been identified. A third omega class member was identified and shown via in situ hybridization to exist on chromosome 3 (GSTO3p).[80] The lack of introns and representation in the express sequence tag database led researchers to believe that GSTO3p is a pseudogene.[80] Further investigation of GSTO3p is merited by the fact that the site of the pseudogene corresponds to a region that was previously believed to contain a gene that may influence the age of onset for Alzheimer’s and Parkinson’s disease.[80]
4.4. Pi Class
A single gene located on chromosome 11 encodes for proteins designated in the Pi class (GSTP1). The GSTP1 gene spans ~3kb, encodes 210 amino acids in seven exons.[81] Expression of GSTP1 has been identified in all tissues and cells, except red blood cells.[82] GSTP1 has been of particular interest because it is overexpressed in a wide variety of tumors.[83,84] The allele frequencies for GSTP1 *A, *B, and *C in Caucasian populations are 0.685, 0.262, and 0.068, respectively.[85] The promoter region contains a TATA box, two SP1 sites, an insulin response element and an antioxidant response element within an AP1 site.[86]
Numerous studies have been published showing the expression of GSTP1 is associated with clinical outcome in cancer (table V).[52,87,88] Polymorphisms at the GSTP1 locus result in four alleles, GSTP1*A-D, that differ structurally and functionally.[86,89] Normal lung tissue from 34 patients was genotyped and analyzed for GST enzyme activity as measured by 1-chloro-2,4-dinitrobenzene conjugation.[90] In these studies, enzyme activity was reduced in individuals expressing one of the GSTP1 Val105 alleles (*B and *C) compared with individuals containing the Ile105 alleles, (*A and *D) [see table III].[90] In separate studies, the Val105 and Ile105 alleles were analyzed to determine if the genotype was a risk factor for a subgroup of basal cell carcinoma patients who develop multiple tumors.[91] GSTP1 Val105/Val105 genotype (BB, BC, or CC) was associated with an increased number of tumors compared with the Ile105/Ile105 genotype (AA, AD, or DD).[91] Perhaps this reflects some degree of susceptibility differences to carcinogen exposure as a consequence of different detoxification profiles for these isozymes.
Table V.
| Allele | Frequency in Caucasian populations | Cancer risks |
|---|---|---|
| P1*A | 0.65 | Cisplatin resistance |
| P1*B | 0.262 | Favorable response to cisplatin. Increased susceptibility to lung, bladder and testicular tumors |
| P1*C | 0.068 | Predominant genotype in malignant glioma. Protective for breast cancer |
The GSTP1 genotype has been associated with differences in cancer and respiratory disorder susceptibilities and response to chemotherapeutic agents. For example, GSTP1*A has been reported to play a role in the acquisition of resistance to cisplatin via formation of platinum-glutathione conjugates.[92] GSTP1*B is an allele in which a single nucleotide (A→G) substitution at position 313 results in the Ile→Val substitution that substantially reduces catalytic activity.[90] Individuals expressing the Val313 allele have a diminished detoxification capacity.[97] Homozygosity for GSTP1*B is favorable in the treatment of patients with cancer because such patients have a diminished capacity to detoxify platinum-based anticancer agents.[93] However, this phenotype is also associated with an increased susceptibility to lung, bladder, and testicular cancers.[94,95] GSTP1*C, an allelic variant that is predominant in malignant glioma cells, differs from other GSTP1 variants by two transitions resulting in Ile105Val and Ala113Val.[86] The GSTP1*C was shown to be protective against breast cancer.[96] However, the precise relevance of this variant to disease occurrence or progression is not yet clear.
4.5. Theta Class
Two genes separated by 50kb on chromosome 22 encode for proteins designated in the Theta class of GSTs, GSTT1 and GSTT2.[98–100] Polymorphisms exist within both genes. GSTT1*A and GSTT1*B differ by a single nucleotide substitution that alters the amino acid residue 104 from threonine (GSTT1*A) to proline (GSTT1*B) [see table III].[101] Introduction of a proline in this region containing an alpha helix results in a conformational change that significantly decreases the activity and mimics the null phenotype. In Swedes, the allele frequency for GSTT1*A is 0.65 versus 0.35 for the non-functional GSTT1*B allele.[101] A deletion in the GSTT1 locus (GSTT1*0) results in a null phenotype in which individuals do not express catalytically active protein. Occurrence of the null phenotype varies between ethnic groups and is found to be highest in Chinese (64.4%) and lowest in Mexican Americans (9%).[102] The null phenotype is also associated with an increased risk for tumors of the head and neck, oral cavity, pharynx, and larynx.[59,88] Countless studies have been published addressing the role of GSTT1 polymorphisms and clinical outcome.[52,53,103]
A new allele has recently been identified in GSTT2 that has a rare amino acid substitution (Met139 Ile).[98] While the frequency of this allele is more prominent in Australian and European populations, any possible phenotype has yet to be identified.
4.6. Zeta Class
A single 10.9kb gene located on chromosome 14 encodes the protein designated inthe Zeta class (GSTZ1).[104,105] GSTZ1 catalyzes the GSH-dependent transformation of a variety of alpha-halogenated acids. GSTZ1 was independently characterized and described as maleylacetoacetate isomerase (MAAI) because it plays a putative role as an isomerase in the catabolic pathway of phenylalanine and tyrosine.[106] GSTZ1, a 29 kDa protein, is expressed in hepatocytes and proximal convoluted tubules. Polymorphisms within the GSTZ1 gene have been identified, and are designated GSTZ1*A–D.[105,107,108] The GSTZ1*A isozyme has been shown to have the highest catalytic activity toward dichloroacetic acid and is predicted to play a key role in the treatment of lactic acidosis where dichloroacetic acid is prescribed.[109] Indeed, repeated treatments with dichloroacetic acid increases its plasma elimination half-life, suggesting that it may induce GSTZ activity.[110] In contrast, GSTZ1*D, characterized by a Thr82Met substitution, has a lower catalytic activity with dichloroacetic acid.[107] This isozyme has been associated with tyrosine metabolism. However, while inborn errors in tyrosine metabolism have been attributed to mutations in other enzymes, none have so far been associated with GSTZ1.[106] Nonetheless, deficiency of GSTZ1 expression has been identified in four families and is associated with mortality within the first year of life.[106] While the precise role (if any) of GSTZ1*D in contributing to premature death remains unclear, it remains plausible that a perturbation in tyrosine metabolism may be contributory.
4.7. The GST Null Phenotype
The Theta and Mu GST classes contain gene deletions that result in a ‘null’ genotype and the absence of enzyme expression and activity. However, a ‘null’ phenotype is observed in individuals with alleles that have a decreased rate of detoxification. GSTM3, GSTP1, and GSTZ1 provide examples of polymorphisms with decreased enzymatic activity. These individuals (homozygous GSTM3*B, GSTP1*B and *A, and GSTZ1*C and *D) are thought to be at risk of a higher level of carcinogen induced damage and, therefore, at higher risk of developing cancers.
Several studies have investigated the expression of GSTs as a unpredictive factor for treatment outcome and survival in patients with cancer.[52,53,103] The results have been varied based on tumor type and progression. In 81 women with invasive ovarian cancer, patients with a null phenotype for GSTM1 or GSTT1 had a better survival after chemotherapy than other patients.[111] In contrast, 148 women with epithelial ovarian cancer who had a null genotype for GSTM1 or GSTT1 showed a poorer prognosis and decrease in disease free interval as compared with women who had GSTM1 or GSTT1 activity.[57]
GSTM1 and GSTT1 genotypes were examined in children with acute myeloid leukemia (AML). These studies showed that individuals who lacked GSTT1 expression (GSTT1*0) displayed a greater toxicity and reduced survival following chemotherapy.[112] In contrast, the null genotype for GSTM1 and GSTT1 conferred a two-fold reduced risk of relapse in children with ALL (see section 4.2).[113] Collectively these studies suggest that the mechanisms that determine survival and disease free interval remain unclear, however, GSTs appear to have a contributory role that varies contingent upon tumor type.
A null genotype for GSTP1 has not been reported. However, hypermethylation of the GSTP1*A regulatory region is the most common somatic alteration identified in human prostate cancer.[114] This alteration results in the loss of GSTP1 expression and is proposed to occur during pathogenesis of the disease.[115] The impact of this phenotype has been extensively studied.[116] Recently, a methyl-CpG binding domain (MBD) protein has been identified that mediates hypermethylation of the GSTP1*A regulatory region.[117] These findings provide a possible target for restoration of GSTP1*A activity. GST expression (and/or activity) of specific isoforms is lost in some individuals with allelic variation. Although it has been speculated that reduced detoxification of possible carcinogens may be causal to malignant transformation and disease progression, a more plausible link may be through an altered capacity to regulate kinase-dependent proliferation pathways.
5. Proteomic Considerations
Evidence for post-translational modifications of GSTs does exist. However, the interpretation of their biological relevance is hampered by the consideration that the experimental evidence is in vitro in nature. For example, GSTP1*A may be subject to phosphorylation at Thr109, Ser28, Ser154 and Ser184; O-glycosylation at Thr5; methylation at unknown sites; N-glycosylation at unknown sites. While the phosphorylation and O-glycosylation are based on predictive sequence modeling, there is direct experimental evidence (albeit with purified proteins) for the methylation[118,119] and N-glycosylation.[120] Whether or not these modifications are species-specific or exist in a cellular milieu, and whether they influence GST function, remains to be established.
6. Pharmacogenetics of GSTs
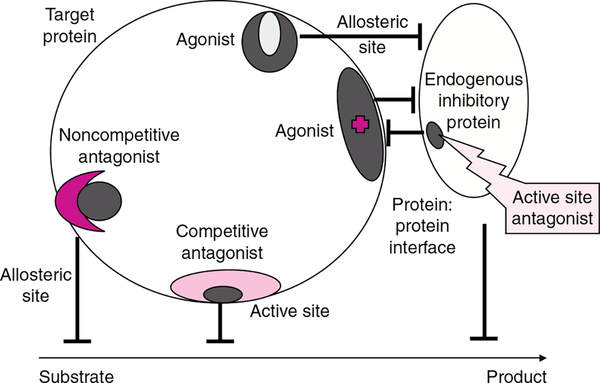
As is apparent from the foregoing discussion, a number of interrelated factors contribute to making GSTs a viable target for cancer drug design. In tumors where GST over-expression imparts a decreased therapeutic response, distinct strategies have been adopted to target GSTs. The first involves the design of GST inhibitors to sensitize tumors where conventional anticancer agents are subject to catalytic detoxification by GSTs. Somewhat novel in concept, progress has been made in the design of compounds to disrupt the protein:protein interactions of GST with interacting proteins, exemplified by stress kinases. This approach, in particular, takes advantage of an emerging principle in drug discovery. While the historical method of discovering new agents has relied upon the targeting of a specific protein to alter its function, it is now possible to target interacting proteins causing interference, with the expectation of a resultant pharmacological effect (see figure 1). Perhaps the most logical of the design strategies is to exploit the elevated expression of GST in tumors, particularly GSTP1*A, through design of GST-activated prodrugs.
Fig. 1.
Drug discovery has frequently involved the targeting of a specific protein with small molecules that act as antagonists or agonists either at active or allosteric sites. Whether resultant drugs act competitively or non-competitively depends upon the actual site and avidity of binding. The impact on a target enzyme is usually interference with substrate-to-product conversion. More recently, the principle of interference with protein:protein interaction as a viable drug discovery approach has gained credibility. Shown in the figure is the concept of targeting an endogenous inhibitory protein (in context of this review, for example GSTP1*A). The outcome is to disassociate this protein from its partner (e.g. c-Jun N-terminal kinase) producing the agonist effect and concomitant activation of the kinase.
6.1. Modulating Drug Resistance in Tumors by GST Inhibition
6.1.1. GST Inhibition by Ethacryinic Acid
A variety of GST inhibitors have been shown to modulate drug resistance by sensitizing tumor cells to anticancer drugs.[121–123] The first clinical studies tested an FDA-approved drug, ethacrynic acid (EA). EA acts non-specifically to inhibit GST Alpha, Mu, and Pi class isozymes by binding directly to GSTs, as well as to deplete its cofactor, GSH, via EA-GSH conjugation, where the thioether conjugate is also an inhibitor of the enzyme.[124,125] EA has been reported to potentiate the cytotoxic effects of alkylating agents, including chlorambucil in human colon carcinoma cell lines and melphalan in human colon tumor xenografts in SCID mice.[121,126]
As a chemosensitizer, the therapeutic value of EA has been demonstrated in patients. A phase I clinical trial showed that EA could suppress GST activity by approximately 50% in white blood cells. This could be correlated with preclinical data showing a corresponding two- to three-fold increase in sensitivity to alkylating agents.[121] However, the efficacy of EA in the clinical management of patients with cancer was limited by a lack of isozyme specificity and its dose-limiting diuretic properties.[127] Clinical correlates to laboratory-based observations are not easily demonstrated. However, in a population of patients with chronic lymphocytic leukemia (CLL), an elevated level of GST activity was found in patients who had received significant treatment with chlorambucil/corticosteroid combinations.[128] A small scale clinical study showed that these same patients, who had developed resistance to therapy, were able to achieve further remission when chlorambucil was administered in combination with EA.[129]
6.1.2. GST Inhibition by TLK199
Efforts to develop inhibitors continued, with improved isozyme specificity and superior clinical application. One such lead compound, TLK199, is a glutathione analog that is a selective inhibitor of GSTP1*A.[130,131] TLK199 acts as a chemosensitizer and was shown to potentiate the toxicity of numerous anticancer agents in different tumor cell lines. In the same study, sensitivity to melphalan was enhanced in xenograft models with elevated GST levels.[132] TLK199 has also been shown to be an effective micromolar inhibitor of the multidrug resistance-associated protein1 (MRP-1), achieving a reversal in the resistance of a variety of agents in NIH3T3 cells transfected with MRP-1.[133]
A serendipitous outcome to the preclinical studies with TLK199 was the observation that TLK199 behaves as a small molecule myeloproliferative agent in rodents.[134] A plausible mechanism for the myelostimulatory effects of TLK199 may be the previously discussed capacity of the drug to disrupt protein: protein interactions in the GSTP:JNK complex. As a consequence of this effect, JNK activity is enhanced and this could be causally associated with the mitogenic response in bone marrow progenitor cells. Elevated levels of JNK have been identified in HL60 cells chronically exposed to, and grown in the presence of, TLK199.[134] This was distinct from the parental (wild-type) cell line where drug exposure induced apoptosis. In vivo studies in myelosuppressed rodents showed a dose-dependent increase in peripheral platelet and neutrophil counts within 24 hours of treatment with a physiological concentration of TLK199.[132,135]
6.2. Modulating Antimicrobial Drug Response by GST Inhibition
GSTs may also be viable drug targets in disease states unrelated to cancer. For example, GSTs distantly related to the mammalian counterparts are present in parasitic organisms and can provide potential targets for therapeutic intervention. Because of the restricted homology, there is always the possibility of an enhanced therapeutic index, since targeting the parasitic protein might have the potential advantage of not compromising the human host. Many anti-parasitic drugs form free radicals that may be inactivated by GSTs from the parasite.[136] In particular, chloroquinone, an antimalarial agent is inactivated by GSH conjugation.[137] The GSH depletion and GST inhibition can result in an enhanced efficacy of chloroquinone against the malarial parasite. Hence, inhibition of parasitic GSTs or destabilization of intraparasitic pools of GSH give a duality of function to therapy with chloroquinone.[137,138] To discover the next generation of anti-malarial drugs, structure-based drug design investigations are underway utilizing crystal structures of the malarial parasites’ GST.[138]
Schistosomiasis, a debilitating tropical disease caused by the parasite Schistosoma japonicum, affects over 200 million people world-wide and results in about 500 000 deaths annually.[138,139] Present therapy for the disease uses oltipraz, a drug which binds directly to the schistosome GST in the integument of the trematode.[140] Development of an effective vaccine is a viable goal for long-term prevention. In fact, a variety of schistosome target antigens are capable of protecting experimental animals from challenge. One of these is a potential vaccine, the 28 kDa S. mansoni GST (Sm28GST) that confers protective immunity in transgenic mice expressing Sm28GST.[48,140] Vaccination with Sm28GST was shown to decrease parasite fecundity and effect egg maturation, thereby decreasing disease pathology in host rats, mice and baboons. Following vaccination in human populations, an inverse correlation was found between IgA antibody production to Sm28GST and a decrease in parasitic egg production.[139] An alternative vaccine directed against the S. haematobium GST (Sh28GST) was shown to be well tolerated in phase I and II clinical trials and demonstrated the capacity to block parasite transmission.[141]
6.3. GSTP1-Activated Prodrugs
Traditional cancer drugs are cytotoxins that target rapidly dividing cells. In most cases, the therapeutic index is compromised because normal tissues, such as bone marrow, gut mucosa, and hair follicles receive exposure equivalent to the tumor. In an effort to improve drug efficacy, greater tumor targeting is a desirable endpoint. Prodrugs are rationally designed inactive agents that are converted to active cytotoxic agents that can target tumor tissues with high expression of activating enzymes. This strategy allows for an increased delivery of active agent to the tumor tissue while minimizing the toxicity towards normal tissues. GSTs provide a promising target because expression is enhanced in many tumors and high levels are sometimes correlated with poor prognosis. In addition, GSTP1*A is frequently elevated in drug resistant tumors. Thus, a two-pronged attack may be afforded by such an activation strategy.
One such approach has been to attempt to exploit the ability of GSTs to catalyze GSH conjugation. For example, cis-3-(9H-purin-6-ylthio)acrylic acid (PTA) is a prodrug of the antitumor and immunosuppressive antimetabolite 6-mercaptopurine that requires GSH conjugation and subsequent metabolism for activation.[142] Renal and hepatic GSTs enhance activation nearly two-fold both in vivo and in vitro as compared with spontaneous GSH conjugation.[142] At this time, this drug is in early preclinical development. However, future success may be restricted by the somewhat narrow spectrum and limited efficacy of the parent drug 6-mercaptopurine.
A second approach in drug development has been to design prodrugs that are selectively activated by GSTs, which are known to be over-expressed in a wide variety of tumors. This design exploits GSTs ability to mediate cleavage of sulfonamides by promoting a β-elimination reaction. Synthesis of drugs as inactive compounds via GSH conjugation through a sulfone linkage was a logical extension of this concept. One such drug is a GSH analog of cyclophosphamide that was shown to selectively enhance toxicity via GST activation in cell and animal models.[143]
TLK286, γ-glutamyl-α-amino-β-(2-ethyl-N,N,N′,N′-tetrakis [2-chloroethyl]phosphorodiamidate)-sulfonyl-propionyl-(R)-(−) phenylglycine, is the lead candidate from a novel class of prodrugs activated in cancer cells by GSTP1*A.[131] GSTP1*A promotes a β-elimination reaction that cleaves TLK286 into a GSH analog and a nitrogen mustard that can alkylate cellular nucleophiles.[144] Sensitivity to the drug is correlated with GSTP1*A expression both in cell culture and in animal models.[144,145] In contrast, lower expression of GSTP1*A at both the protein and transcript level was reported as an adaptive survival trait following chronic exposure to TLK286.[145] These results support the concept that tumors expressing high levels of GSTP1*A will be more sensitive to the cytotoxic effects of TLK286.
The efficacy of TLK286 was examined both in vitro and in vivo. Clonogenic assays showed TLK286 had significant activity against 15 of 21 lung tumors and 11 of 20 breast tumors.[146] Reports on phase I clinical trials showed minor drug related adverse effects that included hematuria and myelosuppression, combined with anti-tumor activity and/or disease stabilization in patients with various advanced malignancies.[147,148] None of the toxicities were greater than grade III, with no indications to limit further clinical development. Clinical benefit has now been reported in phase II trials underway in patients with advanced non-small cell lung cancer (NSCLC) and platinum-resistant ovarian cancer.[149] Such results have encouraged the formulation of phase III trials, which will initially focus on combinations of TLK286 and docetaxel (taxotere) in the treatment of NSCLC.
A general conclusion from the initial foray into targeting GSTs is that the approach is viable. Whether the drugs presently traversing the preclinical and clinical testing protocols will reach eventual widespread utility remains to be determined. However, at this point in development the results are encouraging.
Acknowledgements
The authors have provided no information on sources of funding or on conflicts of interest directly relevant to the content of this review.
References
- 1.Wang AL, Tew KD. Increased glutathione-S-transferase activity in a cell line with acquired resistance to nitrogen mustards. Cancer Treat Rep 1985; 69: 677–82 [PubMed] [Google Scholar]
- 2.Tew KD. Glutathione-associated enzymes in anticancer drug resistance. Cancer Res 1994. Aug 15; 54(16): 4313–20 [PubMed] [Google Scholar]
- 3.Tew KD, Monks A, Barone L, et al. Glutathione-associated enzymes in the human cell lines of the National Cancer Institute Drug Screening Program. Mol Pharmacol 1996. Jul; 50(1): 149–59 [PubMed] [Google Scholar]
- 4.Fan M, Chambers TC. Role of mitogen-activated protein kinases in the response of tumor cells to chemotherapy. Drug Resist Updat 2001. Aug; 4(4): 253–67 [DOI] [PubMed] [Google Scholar]
- 5.Litwack G, Ketterer B, Arias IM. Ligandin: a hepatic protein which binds steroids, bilirubin, carcinogens and a number of exogenous organic anions. Nature 1971; 234: 466–7 [DOI] [PubMed] [Google Scholar]
- 6.Cho SG, Lee YH, Park HS, et al. Glutathione S-transferase mu modulates the stress-activated signals by suppressing apoptosis signal-regulating kinase 1. J Biol Chem 2001. Apr 20; 276(16): 12749–55 [DOI] [PubMed] [Google Scholar]
- 7.Saitoh M, Nishitoh H, Fujii M, et al. Mammalian thioredoxin is a direct inhibitor of apoptosis signal-regulating kinase (ASK) 1. EMBO J 1998; 17: 2596–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ichijo H, Nishida E, Irie K, et al. Induction of apoptosis by ASK1, a mammalian MAPKKK that activates SAPK/JNK and p38 signaling pathways. Science 1997. Jan 3; 275(5296): 90–4 [DOI] [PubMed] [Google Scholar]
- 9.Dorion S, Lambert H, Landry J. Activation of the p38 signaling pathway by heat shock involves the dissociation of glutathione-S-transferase mu from ASK1. J Biol Chem 2002; 277: 30792–7 [DOI] [PubMed] [Google Scholar]
- 10.Davis RJ. Signal transduction by the JNK group of MAP kinases. Cell 2000. Oct 13; 103(2): 239–52 [DOI] [PubMed] [Google Scholar]
- 11.Adler V, Yin Z, Fuchs SY, et al. Regulation of JNK signaling by GSTp. EMBO J 1999; 18(5): 1321–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yin Z, Ivanov V, Habelhah H, et al. Glutathione S-transferase p elicits protection against hydrogen peroxide-induced cell death via coordinated regulation of stress kinases. Cancer Res 2000; 60: 4053–7 [PubMed] [Google Scholar]
- 13.Wang T, Arifoglu P, Ronai Z, et al. Glutathione S-transferase P1–1 (GSTP1–1 inhibits c-Jun NH2 terminal kinase (JNK1) signaling through interaction with the carboxyl terminus. J Biol Chem 2001; 276: 20999–1003 [DOI] [PubMed] [Google Scholar]
- 14.Jakobsson PJ, Thoren S, Morgenstern R, et al. Identification of human prostaglandin E synthase: a microsomal, glutathione-dependent, inducible enzyme, constituting a potential novel drug target. Proc Natl Acad Sci U S A 1999. Jun 22; 96(13): 7220–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jakobsson PJ, Morgenstern R, Mancini J, et al. Common structural features of MAPEG: a widespread superfamily of membrane associated proteins with highly divergent functions in eicosanoid and glutathione metabolism. Protein Sci 1999. Mar; 8(3): 689–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mannervik B, Awasthi YC, Board PG, et al. Nomenclature for human glutathione transferases. Biochem J 1992. Feb 15; 282 (Pt 1): 305–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nguyen T, Liu S, Huang H, et al. Regulation of glutathione S-transferase gene expression by oxidative stress. Series Regulation of gluthatione S-transferase gene expression by oxidative stress. Vol. 133. Dublin: Elsevier Science Ltd, 2001: 7 [Google Scholar]
- 18.Lo H, Ali-Osman F. The human glutathione S-transferase P1 (GSTP1) gene is transactivated by cyclic AMP (cAMP) via a cAMP response element (CRE) proximal to the transcription start site. Series The human glutathione S-transferase P1 (GSTP1) gene is transactivated by cyclic AMP (cAMP) via a cAMP response element (CRE) proximal to the transcription start site. Vol. 133. Dublin: Elseiver Science Ltd, 2001: 320–1 [Google Scholar]
- 19.Desmots F, Rissel M, Loyer P, et al. Immunohistological analysis of glutathione transferase A4 distribution in several human tissues using a specific polyclonal antibody. J Histochem Cytochem 2001. Dec; 49(12): 1573–80 [DOI] [PubMed] [Google Scholar]
- 20.Hayes JD, Pulford DJ. The glutathione S-transferase supergene family: regulation of GST and the contribution of the isoenzymes to cancer chemoprotection and drug resistance. Crit Rev Biochem Mol Biol 1995; 30: 445–600 [DOI] [PubMed] [Google Scholar]
- 21.Mannervik B, Danielson UH. Glutathione transferases: structure and catalytic activity. CRC Crit Rev Biochem 1988; 23(3): 283–337 [DOI] [PubMed] [Google Scholar]
- 22.Armstrong RN. Structure, catalytic mechanism, and evolution of the glutathione transferases. Chem Res Toxicol 1997; 10(1): 2–18 [DOI] [PubMed] [Google Scholar]
- 23.Strange RC, Howie AF, Hume R, et al. The development expression of alpha-, mu-and pi-class glutathione S-transferases in human liver. Biochim Biophys Acta 1989; 993(2–3): 186–90 [DOI] [PubMed] [Google Scholar]
- 24.Sundberg AG, Nilsson R, Appelkvist EL, et al. Immunohistochemical localization of alpha and pi class glutathione transferases in normal human tissues. Pharmacol Toxicol 1993. Apr-May; 72(4–5): 321–31 [DOI] [PubMed] [Google Scholar]
- 25.Mannervik B The isoenzymes of glutathione transferase. Adv Enzymol Relat Areas Mol Biol 1985; 57: 357–417 [DOI] [PubMed] [Google Scholar]
- 26.Knapen MF, Peters WH, Mulder TP, et al. A marker for hepatocellular damage. Lancet 2000. Apr 22; 355(9213): 1463–4 [DOI] [PubMed] [Google Scholar]
- 27.Morel F, Rauch C, Coles B, et al. The human glutathione transferase alpha locus: genomic organization of the gene cluster and functional characterization of the genetic polymorphism in the hGSTA1 promoter. Pharmacogenetics 2002. Jun; 12(4): 277–86 [DOI] [PubMed] [Google Scholar]
- 28.Board PG, Webb GC. Isolation of a cDNA clone and localization of human glutathione S-transferase 2 genes to chromosome band 6p12. Proc Natl Acad Sci U S A 1987. Apr; 84(8): 2377–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Islam MQ, Platz A, Szpirer J, et al. Chromosomal localization of human glutathione transferase genes of classes alpha, mu and pi. Hum Genet 1989. Jul; 82(4): 338–42 [DOI] [PubMed] [Google Scholar]
- 30.Chow NW, Whang-Peng J, Kao-Shan CS, et al. Human glutathione S-transferases: the Ha multigene family encodes products of different but overlapping substrate specificities. J Biol Chem 1988. Sep 15; 263(26): 12797–800 [PubMed] [Google Scholar]
- 31.Rozen F, Nguyen T, Pickett CB. Isolation and characterization of a human glutathione S-transferase Ha1 subunit gene. Arch Biochem Biophys 1992. Feb 1; 292(2): 589–93 [DOI] [PubMed] [Google Scholar]
- 32.Suzuki T, Johnston PN, Board PG. Structure and organization of the human alpha class glutathione S-transferase genes and related pseudogenes. Genomics 1993. Dec; 18(3): 680–6 [DOI] [PubMed] [Google Scholar]
- 33.Bredschneider M, Klein K, Murdter TE, et al. Genetic polymorphisms of glutathione S-transferase A1, the major glutathione S-transferase in human liver: consequences for enzyme expression and busulfan conjugation. Clin Pharmacol Ther 2002. Jun; 71(6): 479–87 [DOI] [PubMed] [Google Scholar]
- 34.Coles B, Nowell SA, MacLeod SL, et al. The role of human glutathione S-transferases (hGSTs) in the detoxification of the food-derived carcinogen metabolite N-acetoxy-PhIP, and the effect of a polymorphism in hGSTA1 on colorectal cancer risk. Mutat Res 2001. Oct 1; 482(1–2): 3–10 [DOI] [PubMed] [Google Scholar]
- 35.Rhoads DM, Zarlengo RP, Tu Chen-Pei D. The basic glutathione S-transferases from human livers are products of separate genes. Biochem Biophys Res Commun 1987; 145: 474–81 [DOI] [PubMed] [Google Scholar]
- 36.Hayes JD, Kerr LA, Cronshaw AD. Evidence of glutathione S-transferases B1 B1 and B2 B2 are the products of separate genes and that their expression in human liver is subject to inter-individual variation. Biochem J 1989; 264: 437–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tetlow N, Liu D, Board P. Polymorphism of human alpha class glutathione transferases. Pharmacogenetics 2001. Oct; 11(7): 609–17 [DOI] [PubMed] [Google Scholar]
- 38.Board PG. Identification of cDNAs encoding two human alpha class glutathione transferases (GSTA3 and GSTA4) and the heterologous expression of GSTA4–4. Biochem J 1998. Mar 1; 330 (Pt 2): 827–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johansson AS, Mannervik B. Human glutathione transferase A3–3, a highly efficient catalyst of double-bond isomerization in the biosynthetic pathway of steroid hormones. J Biol Chem 2001. Aug 31; 276(35): 33061–5 [DOI] [PubMed] [Google Scholar]
- 40.Martinez-Lara E, Siles E, Hernandez R, et al. Glutathione S-transferase isoenzymatic response to aging in rat cerebral cortex and cerebellum. Neurobiol Aging 2003. May-Jun; 24(3): 501–9 [DOI] [PubMed] [Google Scholar]
- 41.Desmots F, Rissel M, Pigeon C, et al. Differential effects of iron overload on GST isoform expression in mouse liver and kidney and correlation between GSTA4 induction and overproduction of free radicles. Free Radic Biol Med 2002. Jan 1; 32(1): 93–101 [DOI] [PubMed] [Google Scholar]
- 42.Kazi S, Ellis EM. Expression of rat liver glutathione-S-transferase GSTA5 in cell lines provides increased resistance to alkylating agents and toxic aldehydes. Chem Biol Interact 2002. May 20; 140(2): 121–35 [DOI] [PubMed] [Google Scholar]
- 43.He NG, Singhal SS, Srivastava SK, et al. Transfection of a 4-hydroxynonenal metabolizing glutathione S-transferase isozyme, mouse GSTA4–4, confers doxorubicin resistance to Chinese hamster ovary cells. Arch Biochem Biophys 1996. Sep 1; 333(1): 214–20 [DOI] [PubMed] [Google Scholar]
- 44.Hayes JD, Pulford DJ, Ellis EM, et al. Regulation of rat glutathione S-transferase A5 by cancer chemopreventive agents: mechanisms of inducible resistance to aflatoxin B1. Chem Biol Interact 1998. Apr 24; 111–112: 51–67 [DOI] [PubMed] [Google Scholar]
- 45.Pulford DJ, Hayes JD. Characterization of the rat glutathione S-transferase Yc2 subunit gene, GSTA5: identification of a putative antioxidant-responsive element in the 5’-flanking region of rat GSTA5 that may mediate chemoprotection against aflatoxin B1. Biochem J 1996. Aug 15; 318 (Pt 1): 75–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pearson WR, Vorachek WR, Xu SJ, et al. Identification of class-mu glutathione transferase genes GSTM1-GSTM5 on human chromosome 1p13. Am J Hum Genet 1993. Jul; 53(1): 220–33 [PMC free article] [PubMed] [Google Scholar]
- 47.DeJong JL, Mohandas T, Tu CP. The human Hb (mu) class glutathione S-transferases are encoded by a dispersed gene family. Biochem Biophys Res Commun 1991. Oct 15; 180(1): 15–22 [DOI] [PubMed] [Google Scholar]
- 48.Xu X, Lemaire C, Grzych JM, et al. Expression of a Schistosoma mansoni 28-kilodalton glutathione S-transferase in the livers of transgenic mice and its effect on parasite infection. Infect Immun 1997. Sep; 65(9): 3867–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Widersten M, Pearson WR, Engstrom A, et al. Heterologous expression of the allelic variant mu-class glutathione transferases mu and psi. Biochem J 1991; 276: 519–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smith G, Stanley LA, Sim E, et al. Metabolic polymorphisms and cancer susceptibility. Cancer Surv 1995; 25: 27–65 [PubMed] [Google Scholar]
- 51.McLellan RA, Oscarson M, Alexandrie AK, et al. Characterization of a human glutathione S-transferase mu cluster containing a duplicated GSTM1 gene that causes ultrarapid enzyme activity. Mol Pharmacol 1997; 52(6): 958–65 [DOI] [PubMed] [Google Scholar]
- 52.Coughlin SS, Hall IJ. Glutathione S-transferase polymorphisms and risk of ovarian cancer: a HuGE review. Genet Med 2002. Jul-Aug; 4(4): 250–7 [DOI] [PubMed] [Google Scholar]
- 53.Hayes JD, Strange RC. Glutathione S-transferase polymorphisms and their biological consequences. Pharmacology 2000. Sep; 61(3): 154–66 [DOI] [PubMed] [Google Scholar]
- 54.Lee KA, Kim SH, Woo HY, et al. Increased frequencies of glutathione S-transferase (GSTM1 and GSTT1) gene deletions in Korean patients with acquired aplastic anemia. Blood 2001. Dec 1; 98(12): 3483–5 [DOI] [PubMed] [Google Scholar]
- 55.Lohmueller KE, Pearce CL, Pike M, et al. Meta-analysis of genetic association studies supports a contribution of common variants to susceptibility to common disease. Nat Genet 2003. Feb; 33(2): 177–82 [DOI] [PubMed] [Google Scholar]
- 56.Ambrosone CB, Sweeney C, Coles BF, et al. Polymorphisms in glutathione S-transferases (GSTM1 and GSTT1) and survival after treatment for breast cancer. Cancer Res 2001; 61: 7130–5 [PubMed] [Google Scholar]
- 57.Howells RE, Redman CW, Dahr KK, et al. Association of glutathione S-transferase GSTM1 and GSTT1 null genotypes with clinical outcome in epithelial ovarian cancer. Clin Cancer Res 1998; 4(10): 2439–45 [PubMed] [Google Scholar]
- 58.Katoh T, Nagata N, Kuroda Y, et al. Glutathione S-transferase M1 (GSTM1) and T1 (GSTT1) genetic polymorphism and susceptibility to gastric and colorectal adenocarcinoma. Carcinogenesis 1996; 17(9): 1855–9 [DOI] [PubMed] [Google Scholar]
- 59.Chenevix-Trench G, Young J, Coggan M, et al. Glutathione S-transferase M1 and T1 polymorphisms: susceptibility to colon cancer and age of onset. Carcinogenesis 1995. Jul; 16(7): 1655–7 [DOI] [PubMed] [Google Scholar]
- 60.Loktionov A, Watson MA, Gunter M, et al. Glutathione-S-transferase gene polymorphisms in colorectal cancer patients: interaction between GSTM1 and GSTM3 allele variants as a risk-modulating factor. Carcinogenesis 2001; 22(7): 1053–60 [DOI] [PubMed] [Google Scholar]
- 61.Beuckmann CT, Fujimori K, Urade Y, et al. Identification of mu-class glutathione transferases M2–2 and M3–3 as cytosolic prostaglandin E synthases in the human brain. Neurochem Res 2000. May; 25(5): 733–8 [DOI] [PubMed] [Google Scholar]
- 62.Baez S, Segura-Aguilar J, Widersten M, et al. Glutathione transferases catalyse the detoxication of oxidized metabolites (o-quinones) of catecholamines and may serve as an antioxidant system preventing degenerative cellular processes. Biochem J 1997. May 15; 324 (Pt 1): 25–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Van Cong N, Laisney V, Gross MS, et al. Glutathione-S-transferases: tissue distribution, number of loci, polymorphism, chromosome localization. Cytogenet Cell Genet 1984; 37: 554 [Google Scholar]
- 64.Campbell E, Takahashi Y, Abramovitz M, et al. A distinct human testis and brain mu-class glutathione S-transferase: molecular cloning and characterization of a form present even in individuals lacking hepatic type mu isoenzymes. J Biol Chem 1990. Jun 5; 265(16): 9188–93 [PubMed] [Google Scholar]
- 65.Inskip A, Elexperu-Camiruasa J, Buxton N, et al. Identification of polymorphism at the gluthathione S-transferase, GSTM3 locus: evidence for linkage with GSTM1 *A. Biochem J 1995; 312: 713–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shi Y, Lee JS, Galvin KM. Everything you have ever wanted to know about Yin Yang 1. Biochim Biophys Acta 1997; 1332(2): F49–66 [DOI] [PubMed] [Google Scholar]
- 67.Yengi L, Inskip A, Gilford J, et al. Polymorphism at the Glutathione S-Transferase Locus GSTM3: interactions with cytochrome P450 and glutathione S-transferase genotypes as risk factors for multiple cutaneous basal cell carcinoma. Cancer Res 1996; 56(9): 1974–7 [PubMed] [Google Scholar]
- 68.Matthias C, Bockmuhl U, Jahnke V, et al. Polymorphism in cytochrome P450 CYP2D6, CYP1A1, CYP2E1 and glutathione S-transferase, GSTM1, GSTM3, GSTT1 and susceptibilty to tobacco-related cancers: studies in upper aerodigestive tract cancers. Pharmacogenetics 1998; 8(2): 91–100 [PubMed] [Google Scholar]
- 69.Hand PA, Inskip A, Gilford J, et al. Allelism at the glutathione S-transferase GSTM3 locus: interactions with GSTM1 and GSTT1 as risk factors for astrocytoma. Carcinogenesis 1996; 17(9): 1919–22 [DOI] [PubMed] [Google Scholar]
- 70.Comstock KE, Johnson KJ, Rifenbery D, et al. Isolation and analysis of the gene and cDNA for a human Mu class glutathione S-transferase, GSTM4. J Biol Chem 1993. Aug 15; 268(23): 16958–65 [PubMed] [Google Scholar]
- 71.Ross VL, Board PG. Molecular cloning and heterologous expression of an alternatively spliced human Mu class glutathione S-transferase transcript. Biochem J 1993. Sep 1; 294 (Pt 2): 373–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liloglou TWM, Laoney P, Youngson J, et al. A T2517C polymorphism in the GSTM4 gene is associated with risk of developing lung cancer. Lung Cancer 2002; 37(2): 143–6 [DOI] [PubMed] [Google Scholar]
- 73.Kearns PR, Chrzanowska-Lightowlers ZM, Pieters R, et al. Mu class glutathione S-transferase mRNA isoform expression in acute lymphoblastic leukaemia. Br J Haematol 2003. Jan; 120(1): 80–8 [DOI] [PubMed] [Google Scholar]
- 74.Board P, Coggan M, Chelvanayagam G, et al. Identification, characterization, and crystal structure of the omega class glutathione transferases. J Biol Chem 2000; 275(32): 24798–806 [DOI] [PubMed] [Google Scholar]
- 75.Adam GC, Sorensen EJ, Cravatt BF. Proteomic profiling of mechanistically distinct enzyme classes using a common chemotype. Nat Biotechnol 2002. Aug; 20(8): 805–9 [DOI] [PubMed] [Google Scholar]
- 76.Washburn MP, Wells WW. Identification of the dehydroascorbic acid reductase and thioltransferase (glutaredoxin) activities of bovine erythrocyte glutathione peroxidase. Biochem Biophys Res Commun 1999; 257: 567–71 [DOI] [PubMed] [Google Scholar]
- 77.Dulhunty A, Gage P, Curtis S, et al. The glutathione transferase structural family includes a nuclear chloride channel and a ryanodine receptor calcium release channel modulator. J Biol Chem 2001; 276(5): 3319–23 [DOI] [PubMed] [Google Scholar]
- 78.Zakharyan R, Sampayo-Reyes A, Healy SM, et al. Human monomethylarsonic acid (MMA (V) reductase is a member of the glutathione-S-transferase super-family. Chem Res Toxicol 2001; 14(8): 1051–7 [DOI] [PubMed] [Google Scholar]
- 79.Tanaka-Kagawa T, Jinno H, Hasegawa T, et al. Functional characterization of two variant human GSTO 1–1s (Ala140Asp and Thr217Asn). Biochem Biophys Res Commun 2003. Feb 7; 301(2): 516–20 [DOI] [PubMed] [Google Scholar]
- 80.Whitbread AK, Tetlow N, Eyre HJ, et al. Characterization of the human Omega class glutathione transferase genes and associated polymorphisms. Pharmacogenetics 2003. Mar; 13(3): 131–44 [DOI] [PubMed] [Google Scholar]
- 81.Cowell IG, Dixon KH, Pemble SE, et al. The structure of the human glutathione S-transferase pi gene. Biochem J 1988. Oct 1; 255(1): 79–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Laisney V, Nguyen Van C, Gross MS, et al. Human genes for glutathione S-transferases. Hum Genet 1984; 68(3): 221–7 [DOI] [PubMed] [Google Scholar]
- 83.Tew KD, Hartley-Asp B. Cytotoxic properties of estramustine unrelated to alkylating and steroid constituents. Urology 1984. Jun; 23(6 Suppl.): 28–33 [DOI] [PubMed] [Google Scholar]
- 84.Moscow JA, Fairchild CR, Madden MJ, et al. Expression of anionic glutathione-S-transferase and P-glycoprotein genes in human tissues and tumors. Cancer Res 1989. Mar 15; 49(6): 1422–8 [PubMed] [Google Scholar]
- 85.Garte S, Gaspari L, Alexandrie A-K, et al. Metabolic gene polymorphism frequencies in control populations. Cancer Epidemiol Biomarkers Prev 2001; 10: 1239–48 [PubMed] [Google Scholar]
- 86.Lo HW, Ali-Osman F. Structure of the human allelic glutathione S-transferase-pi gene variant, hGSTP1 C, cloned from a glioblastoma multiforme cell line. Chem Biol Interact 1998. Apr 24; 111–112: 91–102 [DOI] [PubMed] [Google Scholar]
- 87.Strange RC, Spiteri MA, Ramachandran S, et al. Glutathione-S-transferase family of enzymes. Mutat Res 2001. Oct 1; 482(1–2): 21–6 [DOI] [PubMed] [Google Scholar]
- 88.Strange RC, Fryer AA. The glutathione S-transferases: influence of polymorphism on cancer susceptibility. IARC Sci Publ 1999; 148: 231–49 [PubMed] [Google Scholar]
- 89.Hemmingsen A, Fryer AA, Hepple M, et al. Simultaneous identification of GSTP1 Ile105-->Va1105 and Ala114-->Va11 14 substitutions using an amplification refractory mutation system polymerase chain reaction assay: studies in patients with asthma. Respir Res 2001; 2(4): 255–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Watson MA, Stewart RK, Smith GB, et al. Human glutathione S-transferase P1 polymorphisms: relationship to lung tissue enzyme activity and population frequency distribution. Carcinogenesis 1998. Feb; 19(2): 275–80 [DOI] [PubMed] [Google Scholar]
- 91.Ramachandran S, Hoban PR, Ichii-Jones F, et al. Glutathione S-transferase GSTP1 and cyclin D1 genotypes: association with numbers of basal cell carcinomas in a patient subgroup at high-risk of multiple tumours. Pharmacogenetics 2000. Aug; 10(6): 545–56 [DOI] [PubMed] [Google Scholar]
- 92.Goto S, Iida T, Cho S, et al. Overexpression of glutathione S-transferase pi enhances the adduct formation of cisplatin with glutathione in human cancer cells. Free Radic Res 1999. Dec; 31(6): 549–58 [DOI] [PubMed] [Google Scholar]
- 93.Stoehlmacher J, Park DJ, Zhang W, et al. Association between glutathione S-transferase P1, T1, and M1 genetic polymorphism and survival of patients with metastatic colorectal cancer. J Natl Cancer Inst 2002. Jun 19; 94(12): 936–42 [DOI] [PubMed] [Google Scholar]
- 94.Ryberg D, Skaug V, Hewer A, et al. Genotypes of glutathione transferase M1 and P1 and their significance for lung DNA adduct levels and cancer risk. Carcinogenesis 1997; 18: 1285–9 [DOI] [PubMed] [Google Scholar]
- 95.Harries LW, Stubbins MJ, Forman D, et al. Identification of genetic polymorphisms at the glutathione S-transferase Pi locus and association with susceptibility to bladder, testicular and prostate cancer. Carcinogenesis 1997; 18: 641–4 [DOI] [PubMed] [Google Scholar]
- 96.Maugard CM, Charrier J, Pitard A, et al. Genetic polymorphism at the glutathione S-transferase (GST) P1 locus is a breast cancer risk modifier. Int J Cancer 2001; 91(3): 334–9 [DOI] [PubMed] [Google Scholar]
- 97.Hu X, Xia H, Srivastava SK, et al. Catalytic efficiencies of allelic variants of human glutathione S-transferase P1–1 toward carcinogenic anti-diol epoxides of benzo [c] phenanthrene and benzo [g] chrysene. Cancer Res 1998; 58(23): 5340–3 [PubMed] [Google Scholar]
- 98.Coggan M, Whitbread L, Whittington A, et al. Structure and organization of the human theta-class glutathione S-transferase and D-dopachrome tautomerase gene complex. Biochem J 1998. Sep 15; 334 (Pt 3): 617–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tan KL, Webb GC, Baker RT, et al. Molecular cloning of a cDNA and chromosomal localization of a human theta-class glutathione S-transferase gene (GSTT2) to chromosome 22. Genomics 1995; 25(2): 381–7 [DOI] [PubMed] [Google Scholar]
- 100.Pemble S, Schroeder KR, Spencer SR, et al. Human glutathione S-transferase theta (GSTT1): cDNA cloning and the characterization of a genetic polymorphism. Biochem J 1994; 300 (Pt 1): 271–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Alexandrie AK, Rannug A, Juronen E, et al. Detection and characterization of a novel functional polymorphism in the GSTT1 gene. Pharmacogenetics 2002. Nov; 12(8): 613–9 [DOI] [PubMed] [Google Scholar]
- 102.Nelson HH, Wiencke JK, Christiani DC, et al. Ethnic differences in the prevalence of the homozygous deleted genotype of glutathione S-transferase theta. Carcinogenesis 1995. May; 16(5): 1243–5 [DOI] [PubMed] [Google Scholar]
- 103.Rebbeck TR. Molecular epidemiology of the human glutathione S-transferase genotypes GSTM1 and GSTT1 in cancer susceptibility. Cancer Epidemiol Biomarkers Prev 1997. Sep; 6(9): 733–43 [PubMed] [Google Scholar]
- 104.Board PG, Baker RT, Chelvanayagam G, et al. Zeta, a novel class of glutathione transferases in a range of species from plants to humans. Biochem J 1997. Dec 15; 328 (Pt 3): 929–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Blackburn AC, Woollatt E, Sutherland GR, et al. Characterization and chromosome location of the gene GSTZ1 encoding the human Zeta class glutathione transferase and maleylacetoacetate isomerase. Cytogenet Cell Genet 1998; 83(1–2): 109–14 [DOI] [PubMed] [Google Scholar]
- 106.Fernandez-Canon JM, Penalva MA. Characterization of a fungal maleylacetoacetate isomerase gene and identification of its human homologue. J Biol Chem 1998. Jan 2; 273(1): 329–37 [DOI] [PubMed] [Google Scholar]
- 107.Blackburn AC, Coggan M, Tzeng HF, et al. GSTZ1d: a new allele of glutathione transferase zeta and maleylacetoacetate isomerase. Pharmacogenetics 2001. Nov; 11(8): 671–8 [DOI] [PubMed] [Google Scholar]
- 108.Blackburn AC, Tzeng HF, Anders MW, et al. Discovery of a functional polymorphism in human glutathione transferase zeta by expressed sequence tag database analysis. Pharmacogenetics 2000; 10(1): 49–57 [DOI] [PubMed] [Google Scholar]
- 109.Board P, Chelvanayagam G, Jermiin LS, et al. Identification of novel glutathione transferases and polymorphic variants by expressed sequence tag database analysis. Drug Metab Dispos 2001; 29(4): 544–7 [PubMed] [Google Scholar]
- 110.Tzeng HF, Blackburn AC, Board PG, et al. Polymorphism- and species-dependent inactivation of glutathione transferase zeta by dichloroacetate. Chem Res Toxicol 2000. Apr; 13(4): 231–6 [DOI] [PubMed] [Google Scholar]
- 111.Howells RE, Holland T, Dhar KK, et al. Glutathione S-transferase GSTM1 and GSTT1 genotypes in ovarian cancer: association with p53 expression and survival. Int J Gynecol Cancer 2001; 11(2): 107–12 [DOI] [PubMed] [Google Scholar]
- 112.Davies SM, Robison LL, Buckley JD, et al. Glutathione S-transferase polymorphisms and outcome of chemotherapy in childhood acute myeloid leukemia. J Clin Oncol 2001; 19(5): 1279–87 [DOI] [PubMed] [Google Scholar]
- 113.Stanulla M, Schrappe M, Brechlin AM, et al. Polymorphisms within glutathione S-transferase genes (GSTM1, GSTT1, GSTP1) and risk of relapse in childhood B-cell precursor acute lymphoblastic leukemia: a case-control study. Blood 2000; 95(4): 1222–8 [PubMed] [Google Scholar]
- 114.Lin X, Tascilar M, Lee WH, et al. GSTP1 CpG island hypermethylation is responsible for the absence of GSTP1 expression in human prostate cancer cells. Am J Pathol 2001. Nov; 159(5): 1815–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lee WH, Morton RA, Epstein JI, et al. Cytidine methylation of regulatory sequences near the pi-class glutathione S-transferase gene accompanies human prostatic carcinogenesis. Proc Natl Acad Sci U S A 1994. Nov 22; 91(24): 11733–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Jeronimo C, Varzim G, Henrique R, et al. I105V polymorphism and promoter methylation of the GSTP1 gene in prostate adenocarcinoma. Cancer Epidemiol Biomarkers Prev 2002. May; 11(5): 445–50 [PubMed] [Google Scholar]
- 117.Bakker J, Lin X, Nelson WG. Methyl-CpG binding domain protein 2 represses transcription from hypermethylated pi-class glutathione S-transferase gene promoters in hepatocellular carcinoma cells. J Biol Chem 2002. Jun 21; 277(25): 22573–80 [DOI] [PubMed] [Google Scholar]
- 118.Neal T, Wright LS, Siegel FL. Identification of glutathione S-transferase as a substrate and glutathione as an inhibitor of in vitro calmodulin-stimulated protein methylation in rat liver cytosol. Biochem Biophys Res Commun 1988; 156(1): 368–74 [DOI] [PubMed] [Google Scholar]
- 119.Johnson JA, Finn KA, Siegel FL. Tissue distribution of enzymic methylation of glutathione S-transferase and its effects on catalytic activity: methylation of glutathione S-transferase 11–11 inhibits conjugating activity towards 1-chloro-2,4-dinitrobenzene. Biochem J 1992. Feb 15; 282 (Pt 1): 279–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kuzmich S, Vanderveer LA, Tew KD. Evidence for a glycoconjugate form of glutathione S-transferase pi. Int J Pept Protein Res 1991; 37: 565–71 [DOI] [PubMed] [Google Scholar]
- 121.Tew KD, Bomber AM, Hoffman SJ. Ethacrynic acid and piriprost as enhancers of cytotoxicity in drug resistant and sensitive cell lines. Cancer Res 1988. Jul 1; 48(13): 3622–5 [PubMed] [Google Scholar]
- 122.Hall A, Robson CN, Hickson ID, et al. Possible role of inhibition of glutathione S-transferase in the partial reversal of chlorambucil resistance by indomethacin in a Chinese hamster ovary cell line. Cancer Res 1989. Nov 15; 49(22): 6265–8 [PubMed] [Google Scholar]
- 123.Ford JM, Hait WN, Matlin SA, et al. Modulation of resistance to alkylating agents in cancer cell by gossypol enantiomers. Cancer Lett 1991. Jan; 56(1): 85–94 [DOI] [PubMed] [Google Scholar]
- 124.Oakley AJ, Lo Bello M, Mazzetti AP, et al. The glutathione conjugate of ethacrynic acid can bind to human pi class glutathione transferase P1–1 in two different modes. FEBS Lett 1997. Dec 8; 419(1): 32–6 [DOI] [PubMed] [Google Scholar]
- 125.Mulder TP, Peters WH, Wobbes T, et al. Measurement of glutathione S-transferase P1–1 in plasma: pitfalls and significance of screening and follow-up of patients with gastrointestinal carcinoma. Cancer 1997. Sep 1; 80(5): 873–80 [DOI] [PubMed] [Google Scholar]
- 126.Clapper ML, Hoffman SJ, Tew KD. Sensitization of human colon tumor xenografts to L-phenylalanine mustard using ethacrynic acid. J Cell Pharmacol 1990; 1: 71–8 [Google Scholar]
- 127.O’Dwyer PJ, LaCreta F, Nash S, et al. Phase I study of thiotepa in combination with the glutathione transferase inhibitor ethacrynic acid. Cancer Res 1991. Nov 15; 51(22): 6059–65 [PubMed] [Google Scholar]
- 128.Schisselbauer JC, Silber R, Papadopoulos E, et al. Characterization of lymphocyte glutathione S-transferase isozymes in chronic lymphocytic leukemia (CLL). Cancer Res 1990; 50: 3569–73 [PubMed] [Google Scholar]
- 129.Petrini M, Conte A, Caracciolo F, et al. Reversing of chlorambucil resistance by ethacrynic acid in a B-CLL patient. Br J Haematol 1993. Oct; 85(2): 409–10 [DOI] [PubMed] [Google Scholar]
- 130.Lyttle MH, Docker MD, Hui HC, et al. Isozyme-specific glutathione S-transferase inhibitors: design and synthesis. J Med Chem 1994; 37: 189–94 [DOI] [PubMed] [Google Scholar]
- 131.Lyttle MH, Satyam A, Hocker MD, et al. Glutathione-S-transferase activates novel alkylating agents. J Med Chem 1994. May 13; 37(10): 1501–7 [DOI] [PubMed] [Google Scholar]
- 132.Morgan AS, Ciaccio PJ, Tew KD, et al. Isozyme-specific glutathione S-transferase inhibitors potentiate drug sensitivity in cultured human tumor cell lines. Cancer Chemother Pharmacol 1996; 37(4): 363–70 [DOI] [PubMed] [Google Scholar]
- 133.O’Brien ML, Vulevic B, Freer S, et al. Glutathione peptidomimetic drug modulator of multidrug resistance-associated protein. J Pharmacol Exp Ther 1999. Dec; 291(3): 1348–55 [PubMed] [Google Scholar]
- 134.Ruscoe JE, Rosario LA, Wang T, et al. Pharmacologic or genetic manipulation of glutathione S-transferase P1–1 (GSTpi) influences cell proliferation pathways. J Pharmacol Exp Ther 2001. Jul; 298(1): 339–45 [PubMed] [Google Scholar]
- 135.Carver-Moore K, Broxmeyer HE, Luoh SM, et al. Low levels of erythroid and myeloid progenitors in thrombopoietin-and c-mpl-deficient mice. Blood 1996. Aug 1; 88(3): 803–8 [PubMed] [Google Scholar]
- 136.DeCampo R, Moreno SNJ. Free radical intermediates in the anitparasitic action of drugs and phagocytic cells. Free Radic Biol 1984; VI: 243–88 [Google Scholar]
- 137.Davioud-Charvet E, Delarue S, Biot C, et al. A prodrug form of a Plasmodium falciparum glutathione reductase inhibitor conjugated with a 4-anilinoquinoline. J Med Chem 2001. Nov 22; 44(24): 4268–76 [DOI] [PubMed] [Google Scholar]
- 138.Harwaldt P, Rahlfs S, Becker K. Glutathione S-transferase of the malarial parasite Plasmodium falciparum: characterization of a potential drug target. Biol Chem 2002. May; 383(5): 821–30 [DOI] [PubMed] [Google Scholar]
- 139.Capron A, Riveau G, Grzych JM, et al. Development of a vaccine strategy against human and bovine schistosomiasis: background and update. Mem Inst Oswaldo Cruz 1995. Mar-Apr; 90(2): 235–40 [DOI] [PubMed] [Google Scholar]
- 140.Nare B, Smith JM, Prichard RK. Mechanisms of inactivation of Schistosoma mansoni and mammalian glutathione S-transferase activity by the antischistosomal drug oltipraz. Biochem Pharmacol 1992. Mar 17; 43(6): 1345–51 [DOI] [PubMed] [Google Scholar]
- 141.Capron A, Capron M, Dombrowicz D, et al. Vaccine strategies against schistosomiasis: from concepts to clinical trials. Int Arch Allergy Immunol 2001. Jan-Mar; 124(1–3): 9–15 [DOI] [PubMed] [Google Scholar]
- 142.Gunnarsdottir S, Elfarra AA. Glutathione-dependent metabolism of cis-3-(9H-purin-6-ylthio)acrylic acid to yield the chemotherapeutic drug 6-mer-captopurine: evidence for two distinct mechanisms in rats. J Pharmacol Exp Ther 1999. Sep; 290(3): 950–7 [PubMed] [Google Scholar]
- 143.Kauvar KM. GST: targeted candidates. In: Vermeulen NPE, Mulder GJ, Niewenhuyse H, et al. , editors. Glutathione-S-transferases structure, function and clinical implications. London: Taylor & Francis, 1996: 187–98 [Google Scholar]
- 144.Morgan AS, Sanderson PE, Borch RF, et al. Tumor efficacy and bone marrow sparing properties of TER286, a cytotoxin activated by glutathione S-transferase. Cancer Res 1998; 58: 2568–75 [PubMed] [Google Scholar]
- 145.Rosario LA, O’Brien ML, Henderson CJ, et al. Cellular response to a glutathione S-transferase P1–1 activated prodrug. Mol Pharmacol 2000; 58(1): 167–74 [DOI] [PubMed] [Google Scholar]
- 146.Izbicka E, Lawrence R, Cerna C, et al. Activity of TER286 against human tumor colony-forming units. Anticancer Drugs 1997. Apr; 8(4): 345–8 [DOI] [PubMed] [Google Scholar]
- 147.Rosen LS, Brown J, Lux B, et al. Phase I study of TLK286 (glutathione s-transferase P1–1 activated glutathione analogue) in advanced refractory solid malignancies. Clin Cancer Res 2003. May; 9(5): 1628–38 [PubMed] [Google Scholar]
- 148.Henner WD, Figlin RA, Garland LL, et al. Phase 2 study of TLK286 (GST P1–1 activated glutathione analog) in advanced non-small cell lung cancer (NSCLC) [abstract]. 38th Annual Meeting of the American Society of Clinical Oncology; 2002 May 18–21; Orlando (FL) [Google Scholar]
- 149.Kavanagh JJ, Spriggs D, Bookman M, et al. Phase 2 study of TLK286 (GST P1–1 activated glutathione analog) in patients with platinum resistant epithelial ovarian cancer. In: Series phase 2 study of TLK286 (GST P1–1 activated glutathione analog) in patients with platinum resistant epithelial ovarian cancer [abstract 831]. 38th Annual Meeting of the American Society of Clinical Oncology; 2002 May 18–21; Orlando (FL) [Google Scholar]