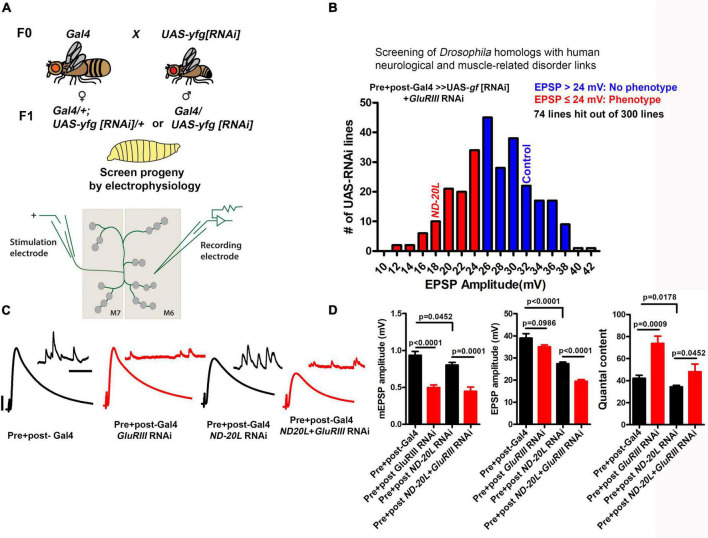
FIGURE 2.
An RNAi screen to identify genes involved in homeostatic synaptic plasticity for human neurological and muscle-related disorders in Drosophila. (A) Crossing scheme for screen. Presynaptic + Postsynaptic-Gal4; GluRIII[RNAi] × UAS-yfg[RNAi] (yfg, “your favorite gene”). For UAS-RNAi lines on chromosomes II or III, male progeny were examined by electrophysiology because dosage compensated elaV(C155)-Gal4/Y male progeny should have a higher dose of pre-synaptic Gal4 than elaV(C155)-Gal4/+ female siblings. (B) The mean value of EPSP amplitudes for screened lines. The mean for the control cross x GluRIII[RNAi] = 35.22 mV (Supplementary Table 1, final row), and standard deviation for this control = 5.88 mV. For our analysis, we have included everything as a potential hit which fell roughly two standard deviations below the control mean amplitude or lower (23.46 mV, ∼24 mV or lower) (Supplementary Table 1). Screen results are highlighted with red and blue bars in the histogram. EPSP cutoffs of ≤ 24 mV (red) were considered putative hits, and > 24 mV (blue) were considered normal (Supplementary Table 1). The majority of knockdown experiments were performed at 29°C, except a few experiments (18°C because higher temperatures caused early lethality). Electrophysiology recordings were carried out at 25°C. (C) Representative electrophysiological traces for selected ND-20L or control genotypes in the presence and absence of GluRIII RNAi. Scale bars for EPSPs (mEPSPs) are x = 50 ms (1000 ms) and y = 10 mV (1 mV). (D) Quantification showing mEPSP, EPSPs amplitude and quantal content in pre + post-Gal4 (mEPSP: 0.93 ± 0.05, EPSP: 38.99 ± 2.02, QC: 42.20 ± 2.65), pre + post-GluRIII RNAi (mEPSP: 0.49 ± 0.03, EPSP: 35.08 ± 0.82, QC: 73.82 ± 6.52), pre + post ND-20L[RNAi] (mEPSP: 0.80 ± 0.03, EPSP: 27.44 ± 0.70, QC: 34.44 ± 1.34) and pre + post ND-20L + GluRIII RNAi (mEPSP: 0.44 ± 0.05, EPSP: 19.55 ± 0.68, QC: 48.18 ± 6.95). The data for pre + post ND-20L[RNAi] and pre + post ND-20L + GluRIII RNAi are also represented in the Supplementary Table 1. Statistical analysis based on one-way ANOVA with Tukey’s post hoc test. Error bars represent mean ± s.e.m. p-values are indicated in the figure.