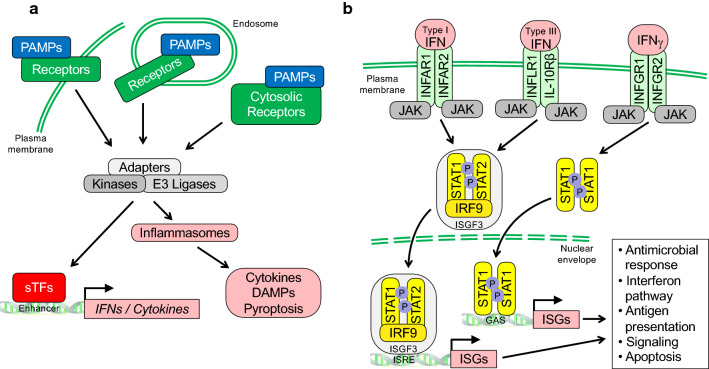
Fig. 1.
Schematic summary of signaling processes in innate immunity. a Pathogen-associated molecular patterns (PAMPs) serve as markers recognized by pattern recognition receptors (PRRs) that allow cells to distinguish between self and non-self. PRRs include membrane bound Toll-like receptors (TLRs) and cytosolic nucleotide-binding oligomerization domain (NOD)-like receptors (NLRs), retinoic acid-inducible gene I (RIG-I)-like receptors (RLRs), AIM2-like receptors (ALRs), and the cyclic guanosine monophosphate–adenosine monophosphate synthase (cGAS). These receptors read different PAMPs, which include conserved microbial components such as glycolipids, peptidoglycans, lipopolysaccharides, and various nucleic acids such as dsRNA and dsDNA, and stimulate signaling complexes that typically involve adaptor proteins and enzymes with kinase and ubiquitin E3 ligase activity. Subsequently, these activate sequence specific transcription factors (sTFs) such as IFN regulatory factors (IRFs) and NF-kB proteins, and inflammasomes. The latter are multimolecular complexes controlling proteolytic enzymes such as caspase-1, which activate IL-1 family cytokines [260, 261]. As a consequence, IFNs, pro-inflammatory cytokines and alarmins or DAMPs (damage associated molecular patterns) are released and disseminate potentially hazardous pathogen encounters. b Different interferons (IFN) interact with distinct heterodimeric receptors as indicated. Upon cytokine binding, Janus family kinases (JAKs) are stimulated that phosphorylate transcription factors of the signal transducer and activator of transcription (STAT) family. Complexes of STAT1 and STAT2 with IRF9 form the trimeric transcription factor ISGF3, which binds to IFN-specific response elements (ISREs). Dimeric STAT1 complexes recognize IFNγ activation sites (GAS). Both ISREs and GAS elements are commonly found in IFN stimulated genes