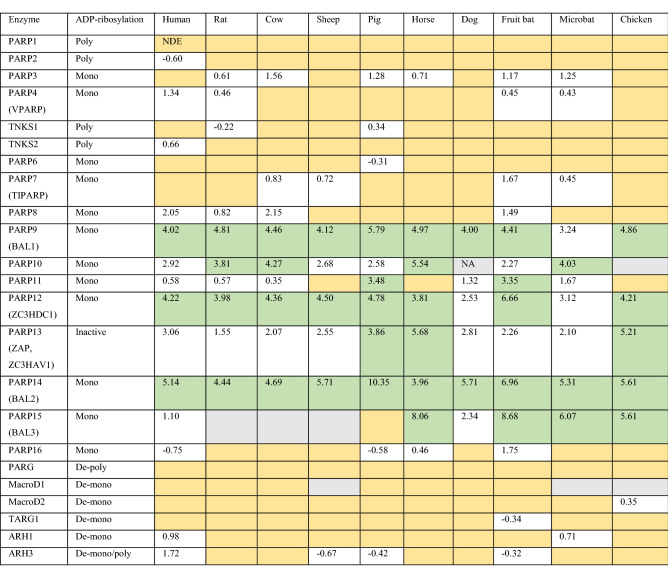
Table 1.
The expression of ARTD subfamily members, macrodomain-containing hydrolases, and ARH family members in response to interferon signaling is summarized
The data were obtained from http://isg.data.cvr.ac.uk [39]
The expression is indicated as log2 fold change. In green are genes whose expression is induced more than tenfold (log2 ≥ 3.33)
NDE/orange: not differentially expressed. This indicates genes that are neither induced nor repressed in a statistically significant manner
NA/grey: no data available
ADP-ribosylation: mono refers to ARTD enzymes capable of mono-ADP-ribosylating substrates; poly to ARTD enzymes synthesizing ADP-ribose polymer chains (iterative mono-ADP-ribosylation); De-poly and de-mono refer to enzymes that can degrade ADP-ribose polymer chains and substrate attached mono-ADP-ribose, respectively