Figure 7: Design based on iGL-CIS43 mice information yields best in class antibody iGL-CIS43.D3.
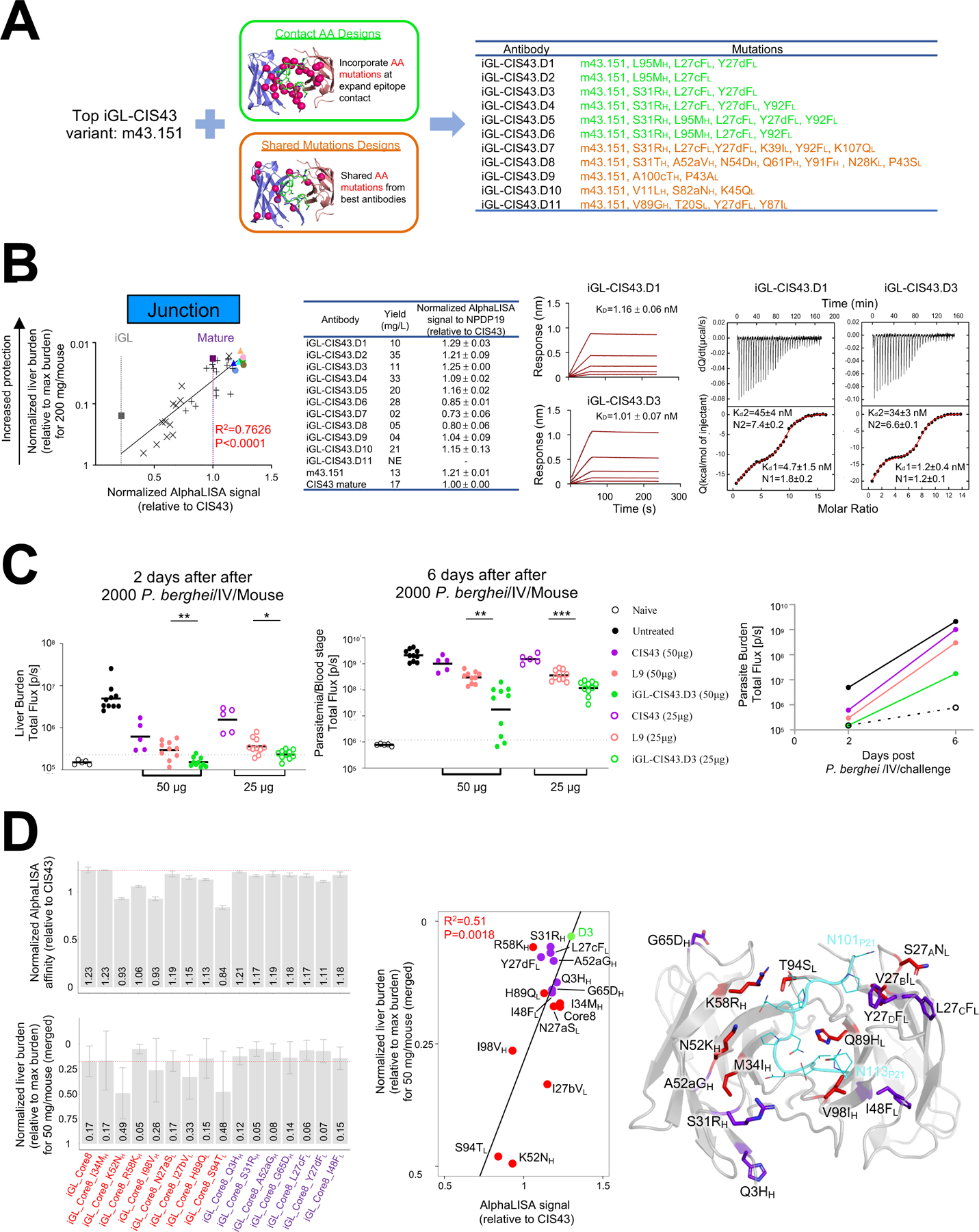
A. m43.151 variants designed using (i) expanded amino acid contact mutations (green color) and (ii) mutations on top antibodies (orange color).
B. AlphaLISA-measured apparent affinity to NPDP19 correlates strongly with protection (R=0.873) for 34 genetically identified antibodies analyzed in Fig. 5 (leftmost panel; antibodies are color coded as in Fig 5C). BLI and ITC affinities for iGL-CIS43.D1 and iGL-CIS43.D3 are shown for NPDP19 and PfCSPm, respectively (right panels).
C. Mice were passively infused with either 50 µg (filed circle) or 25 µg (empty circle) antibody before being challenged with transgenic P. berghei SPZ expressing PfCSPm and a green fluorescent protein/luciferase fusion protein. Bioluminescent quantification was done for each mouse on day 2 and day 6.
D. Structure-function analysis. Initial core 8 mutation were transplanted onto iGL-CIS43 backbone sequence (iGL_Core8) and structure-function analysis was performed, which included the reversion of single mutations and addition of mutations from iGL-CIS43.D3 onto iGL_Core8 construct. AlphaLISA-measured apparent affinities to NPDP19 were determined (D3 has AlphaLISA value 5.53×106), and liver protection was measured (D3 has normalized liver burden 0.03) (left panels). These measurements correlated (middle panel), and the location of each alteration is depicted on the iGL-CIS43.D3 structure (right panel).
