Abstract
The newly discovered SARS-CoV-2 Omicron variant B.1.1.529 is a Variant of Concern (VOC) announced by the World Health Organization (WHO). It's becoming increasingly difficult to keep these variants from spreading over the planet. The fifth wave has begun in several countries because of Omicron variant, and it is posing a threat to human civilization. As a result, we need effective vaccination that can tackle Omicron SARS-CoV-2 variants that are bound to emerge. Therefore, the current study is an initiative to design a peptide-based chimeric vaccine that may potentially battle SARS-CoV-2 Omicron variant. As a result, the most relevant epitopes present in the mutagenic areas of Omicron spike protein were identified using a set of computational tools and immunoinformatic techniques to uncover common MHC-1, MHC-II, and B cell epitopes that may have the ability to influence the host immune mechanism. A final of three epitopes from CD8+ T-cell, CD4+ T-cell epitopes, and B-cell were shortlisted from spike protein, and that are highly antigenic, IFN-γ inducer, as well as overlapping for the construction of twelve vaccine models. As a result, the antigenic epitopes were coupled with a flexible and stable peptide linker, and the adjuvant was added at the N-terminal end to create a unique vaccine candidate. The structure of a 3D vaccine candidate was refined, and its quality was assessed by using web servers. However, the applied immunoinformatic study along with the molecular docking and simulation of 12 modeled vaccines constructs against six distinct HLAs, and TLRs (TLR2, and TLR4) complexes revealed that the V1 construct was non-allergenic, non-toxic, highly immunogenic, antigenic, and most stable. The vaccine candidate's stability was confirmed by molecular dynamics investigations. Finally, we studied the expression of the suggested vaccination using codon optimization and in-silico cloning. The current study proposed V1 Multi-Epitope Vaccine (MEV) as a significant vaccine candidate that may help the scientific community to treat SARS-CoV-2 infections.
Keywords: Reverse vaccinology, COVID-19 omicron variant, Chimeric vaccine model, Spike protein
1. Introduction
In December 2019, a novel Beta coronavirus SARS-CoV-2 was identified as the etiological agent of COVID-19, which has been affecting more than 0.318 billion global populations and resulted in 5.5 million fatalities (https://www.worldometers.info/coronavirus/) (Poon and Peiris, 2020). On the other hand, several variants (i.e. Alpha, Beta, Gamma, Delta, Kappa, and so on) have been evolved as a result of mutations in the SARS-CoV-2 genome since the start of the COVID-19 pandemic. These mutations have a higher impact on the pace of transmission and the immunological escape mechanism (Tao et al., 2021; Thakur et al., 2021). The emergence of multiple variants leads to the emergence of several waves of devastating pandemics around the world (Kumar et al., 2021; Li et al., 2021). Recently, a novel mutant variant of SARS-CoV-2 was identified in Botswana, South Africa. The World Health Organization reported this on November 24, 2021. On November 26, 2021, WHO designated the novel variant Omicron as a Variant of Concern (VOC) (Poudel et al., 2022).
The emergence of a highly mutated SARS-CoV-2 strain (B.1.1.529, Omicron) and its rapid spread across six continents within a week of its first detection has raised worldwide public health concern (Nishiura et al., 2022). Omicron's mutational profile is critical for understanding whether it shares or differs from other SARS-CoV-2 variants in terms of clinical manifestations. Various countries have taken strict actions to minimize the transmission of the Omicron variant of SARS-CoV-2 as a result of the heavily mutated variant emergence. That includes implementing measures, such as restrictions on international flights, strengthened genomic surveillance, and strain sequencing (Petersen et al., 2022).
SARS-CoV-2 pathogenesis is initiated by viral particles adhering to host cell cellular surface receptors, where the Spike glycoprotein (S) of the SARS-CoV-2 virus identifies and binds to Human Angiotensin Converting Enzyme 2 (hACE2) present on the lungs cells (Harrison et al., 2020). Previous studies reported that all the recognized variants showed most of the mutations in their Spike glycoprotein (Harvey et al., 2021). As a result, developing an antiviral drug targeting Spike protein is an appealing strategy for preventing the transmission of COVID variants.
The conventional approach of vaccine development, which involves whole organisms or proteins, results in excessive antigenic load and an increased risk of allergic reactions. This issue can be overcome by developing a Multi-Epitope Vaccine (MEV), which comprises of a short immunogenic peptide, adequate linkers, and an adjuvant capable of eliciting robust and targeted immune responses while reducing allergic reactions. The MEVs are more efficient than single-epitope vaccines because of their specificity towards target, stability of complex, time-saving features, and cost-effectiveness (Bahrami et al., 2019). Furthermore, they are likely to elicit strong humoral and cellular immunological responses at the same time because of the incorporation of T-cell and B-cell epitopes. The MEVs with adjuvants are thought to stimulate long-term immune responses and increased immunogenicity as adjuvants are crucial for reducing the amount of antigen used and the number of shots (Reed et al., 2013). In this continuing pandemic, in silico approaches are the scientifically reliable alternative options for specific vaccine development applicable to all SARS-CoV-2 Variant of Concern (VOC) and Variant of Interest (VOI). Thus, in this study, we hypothesized MEVs against spike protein of Omicron variant using immunoinformatic approaches. In brief, the SARS-CoV-2 Omicron Spike protein was searched for antigenic determinants and then tested to forecast B-cell and T-cell epitopes with Class-I and II MHC (Major Histocompatibility Complex) alleles. Antigenicity, conservancy, and global coverage of anticipated epitopes were investigated. Multiple in silico approaches were also employed to validate the structural stability, physiochemical properties, immunogenicity, toxicity, and antigenicity of the constructed MEVs. The MEV's binding affinity and stability with human pathogenic receptors were further investigated using molecular docking and MD simulations. Finally, the MEV codons were optimized for utilization in the E. coli system, and MEV expression profile was validated using in silico cloning. The findings provide the way for the construction of Omicron variant vaccines, however further experimental validation is needed to battle the pandemic.
2. Material and methods
2.1. Data retrieval
The Omicron spike genomic sequence was obtained from the Global Initiative for Sharing All Influenza Data (GISAID) under the accession number EPI ISL 8616776, which was reported by the National Health Laboratory South Africa, and then translated into amino acids sequence using the BLASTX tool.
2.2. Multiple Sequence Alignment
The spike proteins of Omicron variant and Wuhan (wild type) were aligned to study the variation in these strains of SARS-CoV-2. For this purpose, ClustalO tool was used that work on progressive alignment techniques.
2.3. Reverse Vaccinology
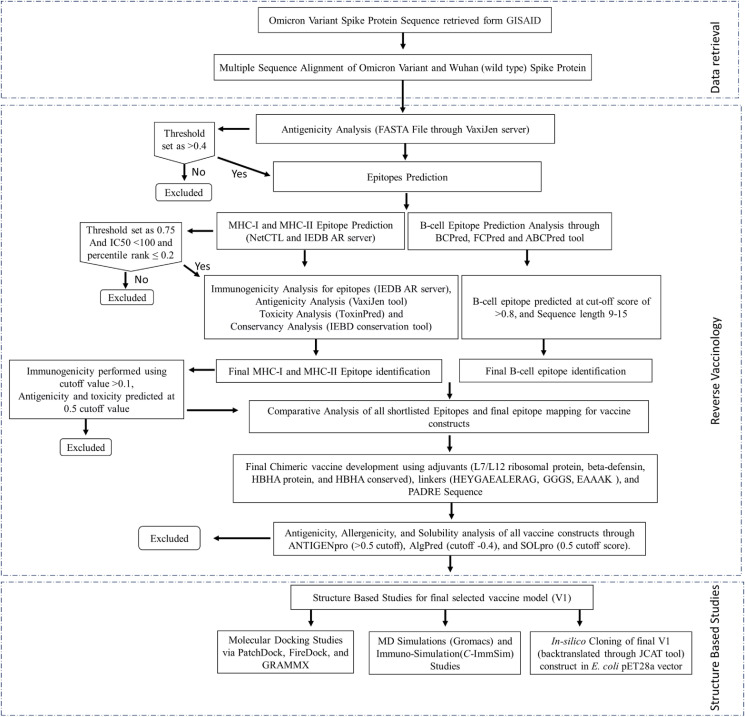
The Vaccinomics, biochemistry, immunology, proteomics, molecular biology, and genomics have all recently evolved, and transformed traditional vaccinology into Reverse Vaccinology (RV). The RV is an innovative and developing computational method that has been widely utilized to improve vaccine target and model prediction especially for those pathogens that are difficult to cultivate in the lab (Moxon et al., 2019). This methodology integrates immunogenomics, immunogenetics, and bioinformatics to scan the whole proteome of a pathogen to find a new vaccine candidate and analyze its ability to elicit a host-immune response. The complete applied reverse vaccinology methodology is highlighted in Fig. 1 .
Fig. 1.
Workflow. The applied study workflow utilized for the construction of potential vaccine model against omicron variant spike protein through reverse vaccinology approach.
2.4. Antigenicity identification
Antigenicity refers to an antigen's capacity of pathogen to be identified by the immune system and elicit an immunological response. The VaxiJen v 2.0 was used to assess the antigenicity of spike protein of Omicron with 0.4 as a threshold value (Doytchinova and Flower, 2007).
2.5. Prediction of MHC-I T-cell epitopes
The immunomodulatory effects of different types of epitopes were studied using the NetCTL service to detect T-cell epitopes from spike protein (Doytchinova et al., 2006; Jalal et al., 2021) using a combined score of anticipated characteristics and a 0.75 threshold (Hasan et al., 2016). The T cell epitopes were also submitted to the Immune Epitope Database and Analysis Resource (IEDB AR) for MHC I binding prediction, which demonstrated that T cells recognized MHC-I antigens (Kim et al., 2012). The IEDB server has lots of information on epitope immunogenicity for adaptive immunity. The default consensus settings of CombLib, ANN, NetMHCpan and SMM together with all HLA alleles and human MHC were used for epitope prediction in the current study (Kim et al., 2012). The HLA alleles analyzed in present study were HLA-A 0205, HLA_0201, HLA-A3, HLA-B 5401, HLA-A2, HLA-A 2.1, and HLA-B 5102 (Almofti et al., 2021). The threshold parameters on the basis of IC50 < 100μM and <0.2 of percentile rank were used as cut-off values for narrowing down of MHC class-I epitopes (Rahman et al., 2020).
2.6. MHC-I immunogenicity, antigenicity, toxicity and conservancy analysis
The T-cell epitope must be immunogenic to trigger either CD4+ or CD8+ T cells. To determine their ability to elicit an immunological response, we employed the IEBD AR tool for MHC-I immunogenicity prediction. The default parameter settings were used for immunogenicity prediction, and positive value epitopes were chosen for future study (Dhanda et al., 2019).
The conservancy, toxicity, and antigenic attributes of promiscuous MHC-I immunogenic epitopes were explored further. The epitope sequence should be conserved across all detected variant sequences in order to construct broad-spectrum peptide-based vaccines (Esmailnia et al., 2020). The IEBD server was utilized to perform the conversancy analysis (Angelo et al., 2017). The antigenicity of conserved epitopes was also predicted using the VaxiJen server, which has a 70–80% accuracy rate and a 0.5 probability threshold score (Doytchinova and Flower, 2007). The relative toxicity level of epitopes was determine through ToxinPred server with cut-off value set as 0.5 (Gupta et al., 2013).
2.7. T-cell MHC-II prediction
A consensus-based technique was applied for epitopes prediction with binding potency to MHC class-II through the IEBD-AR server (Hajialibeigi et al., 2021). In order to shortlist MHC-II epitopes, a cut-off value of 0.2 peptide rank and an IC50 of 100 nM for top binders were determined against the worldwide human population's 95 percent HLA variability, i.e., DRB1*0401, DRB1*0701, DRB301:01, DRB1*1301, DRB1*0101, DRB1*0301, DRB1*0801, DRB1*1101, HLA-DRB401:01, DRB1*1501 (Solanki and Tiwari, 2018). Multiple epitopes with 9–14 residues were chosen for downstream investigation.
2.8. Identified MHC restricted alleles
The identified MHCI – II epitopes were clustered to validate their respected MHC restricted alleles using the MHCcluster (Thomsen et al., 2013). The clustering performed for these epitopes is resulted in the plotting of heat map for the expression relation epitopes with corresponding alleles. It also generated phylogenetic tree that helped in the assessment of functional relation identification between HLAs and shortlisted epitopes (Ullah et al., 2020).
2.9. B-cell epitope identification
A perfect peptide vaccine may induce long-lasting humoral immunity, identical to the natural immunological response produced by pathogenic infections. The B-cell epitopes stimulate humoral immunity, which can kill infection by creating antibodies against exposed antigens to the human body. The ABCPred (Saha and Raghava, 2006), BCPred (Bhattacharya et al., 2019), and FCPred (EL-Manzalawy et al., 2008) servers are using sequence-based technique and a cut-off score of >0.8 was used for the identification of B-cell epitopes. Moreover, ElliPro server (Ponomarenko et al., 2008) was used for the assessment of hydrophobicity content of B-cell epitopes (EL-Manzalawy et al., 2008), Chou and Fashman tool (Chou, 1978) for beta turn prediction, Kaprplus and Schulz for flexibility scale (Karplus and Schulz, 1985), and Parker hydrophilicity scale for the identification of hydrophobic content (Parker et al., 1986), respectively.
2.10. Epitope mapping (B-cell and T-cell)
The development of epitope-based vaccines requires an epitope that can trigger immune cells (B and T cells) to react (Khalid et al., 2022). Consequently, the epitopes having binding affinity and similarity with each other from shortlisted MHC-I/II and B-cell epitopes of Omicron spike protein were determined. Manual comparison was carried out, and overlapping epitopes are classified as plausible peptide sequence. These peptides were compiled and used as final anticipated epitopes for vaccine modeling.
2.11. Vaccines construction and structure modeling
The shortlisted epitopes were conjugated sequentially with suitable adjuvant, PADRE (Pan HLA-DR reactive epitope) sequence, and linkers to create various vaccine constructs. We employed a variety of epitope sequences with four different adjuvants such as L7/L12 ribosomal protein, beta-defensin, HBHA protein, and HBHA conserved sequence to construct a vaccine model with minimal toxicity, allergenicity, and immunogenicity (Y. Yang et al., 2015). The addition of linkers boosts immunogenicity, while the PADRE peptide assists in the production of CD4+ T-cells, improving the efficacy and potency of peptide vaccines (Ghaffari-Nazari et al., 2015). HEYGAEALERAG and GGGS linkers were employed to combine HTL, CTL, and B-cell epitopes, while EAAAK linkers were utilized to link adjuvant sequences at both the N- and C-terminus. The TLR connections that have a profound immuno-stimulatory effect on polarized CTL response were further investigated in the fused vaccine models (Araújo et al., 2019).
2.12. Antigenicity, allergenicity, and solubility assessment for vaccines constructs
The AlgPred server for allergenicity analysis with a cut-off of −0.4 and an accuracy of 85 percent was utilized to examine their allergenicity effects to overcome the allergic reaction characteristic of vaccine constructs. Scores less than a threshold are considered as non-allergenic vaccine (Yukeswaran et al., 2021). The antigenic potential of model vaccines was further predicted through VaxiJen and ANTIGENpro servers with the default threshold value of >0.5. Furthermore, using default settings of 74 percent accuracy and corresponding probability (i.e. 0.5), the SOLpro program was used to determine the solubility characteristic of the vaccine model that will aid in the successful expression of vaccine construct in E. coli plasmid (Dhanda et al., 2019).
2.13. Physiochemical analysis of constructed vaccines
The Expasy ProtParam (Garg et al., 2016) program was used to perform physicochemical analysis and functional characterization of vaccines based on parameters such as pK values, molecular weight, estimated half-life, GRAVY values, aliphatic index, instability index, isoelectric pH and half-life of generated vaccine model (Wang et al., 2020). Physiochemical properties must be assessed to determine the safety and effectiveness of vaccine candidates.
2.14. Structure modeling and molecular dynamic simulation
The interaction of the vaccine component with its receptors was studied to induce a consistent immune response in the vaccine model to target cells. The molecular docking method is a useful tool for estimating the binding energies and assessing interactions between epitopes and HLA molecules (Morris and Lim-Wilby, 2008). The final possible vaccine constructs that met all of the framework's criteria were then docked into the binding cavities of six HLA alleles commonly found in the human population i.e. 1H15 (HLA-DR B5*01:01), 2FSE (HLA-DR B1*01:01), 1A6A (HLA-DR B1*03:01), 3C5J (HLA-DR B3*02:02), 2Q6W (HLA-DR B3*01:01), and 2SEB(HLA-DRB1*04:01) retrieved from the protein database (Burley et al., 2021). The HLAs and vaccine interactions were estimated using the PatchDock server (Schneidman-Duhovny et al., 2005). Furthermore, The GRAMMX tool was also employed to validate the vaccine and TLR4/MD complex docking. The TLR4 is involved in the recognition of viral proteins, which results in the release of inflammatory cytokines. Hu et al. (2012) reported that TLR4 is required for an effective immune response against SARS-CoV-2 (Hu et al., 2012). Therefore, molecular docking of vaccine construct and TLR4 was performed through GRAMMX (Tovchigrechko and Vakser, 2006) and the interactions such as hydrogen bonding were validated and visualized by PDBsum (Laskowski, 2001) and UCSF Chimera (Pettersen et al., 2004). The MD simulation studies were carried out to evaluate the structural stability of the designed vaccine using GROMACS v. 2020 (Van Der Spoel et al., 2005). The topological parameters for the vaccine were processed by using GROMOS96 54A7 force field (Lin and van Gunsteren, 2013). The vaccine was put at 1.0 Å apart from the box edge in a cubic box that was constructed. Further, solvent molecules of SPC water model were added by using periodic boundary conditions. The addition of counter ions neutralized the solvated system. With a maximum force of <1000 kJ mol−1 Å−1, the neutralized system was minimized using the steepest descent technique. The system was gradually heated at constant temperature and pressure in NVT and NPT ensemble for 100 ps using thermostat and Berendsen barostat algorithms. For constraining the bond length and calculation of electrostatic long-range interactions, LINCS algorithm, and PME method was applied, respectively. Finally, a 50 ns MD production was performed with coordinates and energies stored every 10 ps in the output trajectory file, according to established procedure (Jalal et al., 2021). Subsequently, the Root Mean Square Fluctuation (RMSF), Root Mean Square Deviation (RMSD), and Radius of gyration (Rg) were plotted to evaluate the stability of the system. Furthermore, molecular dynamic simulation of docked complex (vaccine with TLR4) was performed via iMODs server. The iMODs defines and estimates the flexibility of protein complex based on the direction and extent of the immanent motions of the complex in terms of deformability, covariance, B-factors, and eigenvalue.
2.15. Immune simulation of final vaccine construct
The immune response profile and vaccine immune simulation were anticipated using the C-ImmSim simulation server (Rahman et al., 2020a, Rahman et al., 2020b). Three injections of modeled prophylactic Omicron variant vaccine were administered at 1, 82, and 126 h time periods and 12345 random seed for up to 4 weeks at three different intervals with default simulation parameters containing no LPS, volume, and simulation stages at 10, and 1000, respectively, with homozygous host haplotypes HLA-A*0101, HLA-A*0201, HLA-B*0702, HLA-DRB1*0101, and HLA-DRB1*0401 (Kaba et al., 2018).
2.16. In-silico cloning and codon optimization of final vaccine construct
The Java Codon Adaptation Tool (JCAT) was used to modify the vaccine's codon within the E. coli host strain k-12 (Grote et al., 2005). The amino acid sequence of the vaccine was back translated to DNA and then modified for codon use in E. coli. The adaptation of vaccine model was expressed in terms of Codon Adaptation Index (CAI) value and GC content estimated through the JCAT algorithm. In addition, the SnapGene tool (Hess et al., 1992) was used to clone the modified gene sequence of the final vaccine construct in E. coli pET28a vector to ensure vaccine construct expression.
3. Results
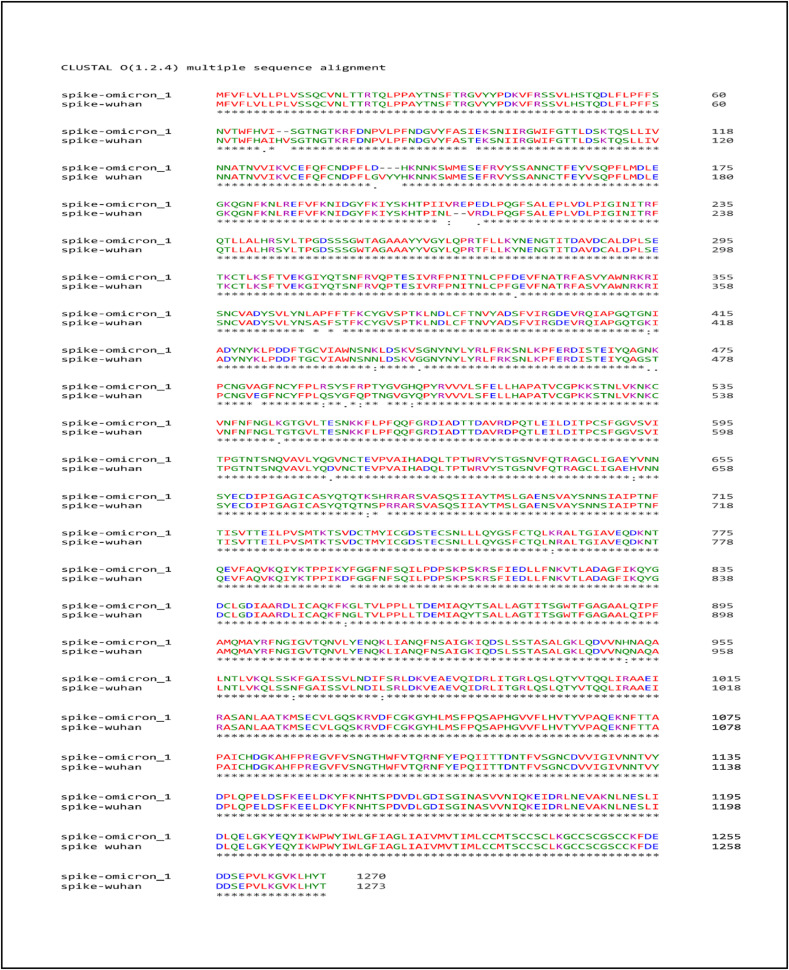
3.1. Multiple Sequence Alignment
The MSA analysis of Omicron spike proteins with Wuhan strain showed 97–100% through identity matrix. It showed that the Omicron spike protein has unique substitutions at 67, 94, 140, 207, 210, 341, 368, 370, 372, 414, 437, 443, 474–475, 481, 490, 493, 495, 498, 502, 544, 611, 652, 675, 760, 853, 951, 966, 978, deletion at position 141–143 and insertion at positions 206–207. The complete visual representation about the mutation detail is provided in Fig. 2 that showed the alignment of spike proteins, change in amino acids, and number of mutations.
Fig. 2.
Multiple Sequence Alignment. Sequence alignment of Omicron spike protein and Wuhan spike protein representing the mutations, insertion, deletion and SNP in Omicron protein.
3.2. Antigenicity prediction for Omicron Spike protein
The antigenicity of Omicron spike proteins analyzed through VaxiJen server was identified as 0.4125 with a cut-off value of 0.4. The analysis indicates the spike protein as antigenic that can stimulate host-immune response.
3.3. Prediction of MHC Class-I T-cell epitopes
The NetCTL server was used to forecast 1261 T-cell epitopes using a threshold value of 0.75 (Supplementary file 1), however, only 131 epitopes were found to exhibit optimum T-cell binding. The MHC-I binding on these 131 epitopes was evaluated using IEBD tool. It identified approximately 2657 MHC-I epitopes (Supplementary file 2). However, based on MHC-I and T-cell interaction, 163 epitopes were identified that evoked strong binding affinity using a cut-off criterion of IC50 < 100 and percentile rank ≤0.2. All the shortlisted epitopes were predicted to be efficient T-cell binders and were evaluated further.
3.4. Immunogenicity antigenicity, conservancy and toxicity analysis for shortlisted epitopes
The immunogenicity of epitopes determines their ability to induce T-cell responses. The immunogenicity prediction was performed for the shortlisted epitopes. The higher the immunogenicity score, the better epitopes will be at stimulating cellular immunity and T-cells. From the 163 MHC-I shortlisted epitopes, 96 immunogenic epitopes with a positive score cut-off were identified as having significant immunogenic values using the IEBD service. These immunogenic selected epitopes were employed in the development of vaccines.
The toxicity, antigenicity, and conservancy analyses were also performed on the 96 immunogenic epitopes that were shortlisted. The results of the ToxinPred and IEBD conservation tools showed that all 96 sequences were non-toxic and 100% conserved. However, antigenicity analysis using the VaxiJen server resulted in a total of 24 epitopes (Table 1 ) as antigenic (i.e., score between 0.5 and 1.0) and subjected to further evaluation whereas discarding the non-antigen one.
Table 1.
Shortlisted MHC-I Epitopes along with their predicted Immunogenicity, Antigenicity, Toxicity, and Conserve Analysis.
| S. No. | Epitopes | Immunogenicity | Antigenicity | Toxicity | Conserve |
|---|---|---|---|---|---|
| 1 | YIKWPWYIW | 0.42524 | 0.9673 | Non-Toxin | 100% |
| 2 | NLAPFFTFK | 0.3269 | 1.3365 | Non-Toxin | 100% |
| 3 | TLADAGFIK | 0.28158 | 0.5781 | Non-Toxin | 100% |
| 4 | YNLAPFFTF | 0.25665 | 0.9319 | Non-Toxin | 100% |
| 5 | QYIKWPWYI | 0.21624 | 1.4177 | Non-Toxin | 100% |
| 6 | IAIPTNFTI | 0.18523 | 0.7052 | Non-Toxin | 100% |
| 7 | IPTNFTISV | 0.17229 | 0.8820 | Non-Toxin | 100% |
| 8 | GVYFASIEK | 0.16979 | 0.4008 | Non-Toxin | 100% |
| 9 | KEIDRLNEV | 0.15852 | 0.5300 | Non-Toxin | 100% |
| 10 | WTAGAAAYY | 0.15259 | 0.6306 | Non-Toxin | 100% |
| 11 | DIADTTDAV | 0.15094 | 1.0904 | Non-Toxin | 100% |
| 12 | FNATRFASV | 0.14872 | 0.5609 | Non-Toxin | 100% |
| 13 | YLQPRTFLL | 0.1305 | 0.4532 | Non-Toxin | 100% |
| 14 | VVFLHVTYV | 0.1278 | 1.5122 | Non-Toxin | 100% |
| 15 | LPFNDGVYF | 0.11767 | 0.5593 | Non-Toxin | 100% |
| 16 | ASANLAATK | 0.08792 | 0.7014 | Non-Toxin | 100% |
| 17 | STQDLFLPF | 0.06828 | 0.6619 | Non-Toxin | 100% |
| 18 | YEQYIKWPW | 0.06574 | 0.8690 | Non-Toxin | 100% |
| 19 | FTISVTTEI | 0.04473 | 0.8535 | Non-Toxin | 100% |
| 20 | PYRVVVLSF | 0.03138 | 1.0281 | Non-Toxin | 100% |
| 21 | VTYVPAQEK | 0.02711 | 0.8132 | Non-Toxin | 100% |
| 22 | RLDKVEAEV | 0.01617 | 0.0765 | Non-Toxin | 100% |
| 23 | RSYSFRPTY | 0.00837 | 0.9553 | Non-Toxin | 100% |
| 24 | AEIRASANL | 0.00689 | 0.7082 | Non-Toxin | 100% |
3.5. MHC-II epitopes identification and conservancy analysis
Furthermore, 916 MHC-II epitopes were identified employing the IEBD server in addition to MHC-I epitope prediction (Supplementary file 3). We evaluated epitopes based on their percentile rank with <0.2 values and a binding affinity with IC50 < 100 nM. A total of 15 MHC-II epitopes were predicted by the server following the applied threshold and 100 percent conserved predicted by the IEBD conservancy analysis for MHC-II epitope (Table 2 ).
Table 2.
Predicted antigenicity, toxicity, and conservancy analysis of shortlisted MHC-II epitopes.
| S. No. | Epitopes | Antigenicity | Toxicity | Conserve |
|---|---|---|---|---|
| 1 | QSLLIVNNATNVVIK | Non-Toxin | 100% | |
| 2 | SLLIVNNATNVVIKV | Non-Toxin | 100% | |
| 3 | FDEVFNATRFASVYA | Non-Toxin | 100% | |
| 4 | TQSLLIVNNATNVVI | Non-Toxin | 100% | |
| 5 | DEVFNATRFASVYAW | Non-Toxin | 100% | |
| 6 | LLIVNNATNVVIKVC | Non-Toxin | 100% | |
| 7 | PFDEVFNATRFASVY | Non-Toxin | 100% | |
| 8 | AQKFKGLTVLPPLLT | Non-Toxin | 100% | |
| 9 | EVFNATRFASVYAWN | Non-Toxin | 100% | |
| 10 | KTQSLLIVNNATNVV | Non-Toxin | 100% | |
| 11 | NCTFEYVSQPFLMDL | Non-Toxin | 100% | |
| 12 | CTFEYVSQPFLMDLE | Non-Toxin | 100% | |
| 13 | QKFKGLTVLPPLLTD | Non-Toxin | 100% | |
| 14 | QQLIRAAEIRASANL | Non-Toxin | 100% | |
| 15 | REGVFVSNGTHWFVT | Non-Toxin | 100% |
3.6. MHC restriction cluster analysis of shortlisted epitopes
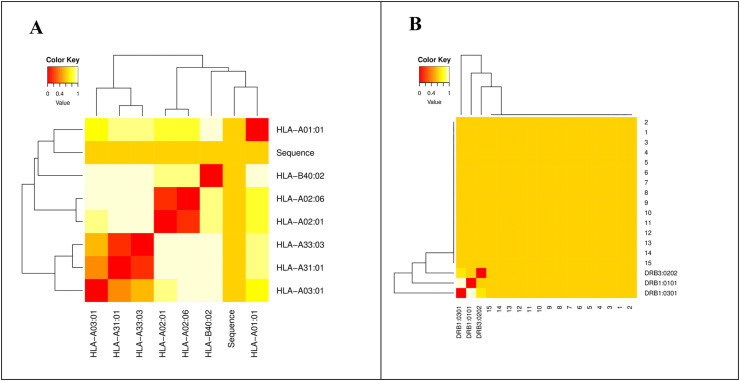
The MHCclusters tool was used to evaluate the identified MHC-I/II epitopes in relation to MHC restricted allele and their suitable peptide validated the epitopes found in T-cells. In terms of annotation, the interaction between MHC-I/II and HLAs is displayed as a heat map and phylogenetic dynamic tree with red color representing stronger interaction and yellow color representing weak interaction (Fig. 3 ).
Fig. 3.
MHCs Restricted Allele analysis. (A) The MHC-I restricted Allele analysis against shortlisted epitopes, while (B) the MHC-II restricted Allele analysis against shortlisted epitopes.
3.7. Prediction of B-cell epitope
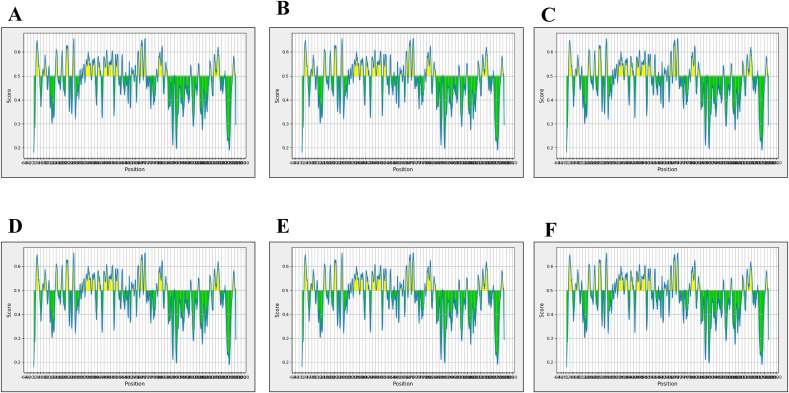
Ideally, both humoral and cellular immunity are required to successfully eradicate the virus from the body. As a result, B-cell epitopes against Omicron spike protein were discovered using ABCPred, BCPred, and FBCPred. These methods identified 155, 22, and 42 B-cell epitopes using a threshold value of 0.51, and a specificity of 75 percent, (Supplementary File 4-Table S1). Chou-Fasman beta-turn prediction, Kolaskar Tongaonkar antigenicity, BepiPred linear epitope prediction, Parker hydrophilicity prediction, Karplus-Schulz flexibility prediction, and Emini surface accessibility prediction were also used to further examine and select the resulting B-cell epitopes as highlighted in Fig. 4 .
Fig. 4.
B-cell epitopes Analysis. (A) Bepipred Linear Epitope, (B) Chou & Fasman Beta-Turn Prediction, (C) Emini Surface Accessibility Prediction, (D) Karplus& Schulz Flexibility Prediction, (E) Kolaskar & Tongaonkar Antigenicity, (F) ParkerHydrophilicity Prediction.
3.8. Epitope mapping and prioritization
The selected B-cell epitopes were used as a template and manually evaluated against MHC-I and MHC-II epitopes to screen out overlapping epitopes. The comparison analysis was resulted in the shortlisting of three epitopes that overlap MHC-I, MHC-II, and B-cell epitopes (Table 3 ) i.e., TESIVRFPNITNLCPFDEVFNATRFASVYAWNRKRISNCVADYSVLYNLAPFFTFKCYG, PQSAPHGVVFLHVTYVPAQEKNFTTAPAICHDGKAHFPREGVFVSNGTHWF, and TQQLIRAAEIRASANLAATKMSECVLGQSKRVDFCGKG, respectively.
Table 3.
Final Epitopes when compared to MHC-I, MHC-II and B-cell Epitopes.
| S. No. | Positions | B-cell Epitopes (Final Epitopes) | MHC-I epitopes | MHC-II Epitopes | Score |
|---|---|---|---|---|---|
| 1 | 320–378 | TESIVRFPNITNLCPFDEVFNATRFASVYAWNRKRISNCVADYSVLYNLAPFFTFKCYG | FNATRFASV | FDEVFNATRFASVYA DEVFNATRFASVYAW PFDEVFNATRFASVY EVFNATRFASVYAWN |
0.74 |
| 2 | 1050–1101 | PQSAPHGVVFLHVTYVPAQEKNFTTAPAICHDGKAHFPREGVFVSNGTHWF | VVFLHVTYV VTYVPAQEK |
REGVFVSNGTHW | 0.65 |
| 3 | 1006–1043 | TQQLIRAAEIRASANLAATKMSECVLGQSKRVDFCGKG | ASANLAATK AEIRASANL |
QQLIRAAEIRASANL | 0.66 |
3.9. Vaccine construction, allergenicity, antigenicity solubility and physicochemical properties prediction
Four adjuvants, PADRE sequences, GGGS, HEYGAEALERAG, and EAAAK linkers were used to insert these four epitopes in a sequential way. The PADRE sequence helped to overcome the universal polymorphism impact of HLA-DR molecules in varied cultures, whereas this combination of adjuvants and linkers leads to activating a large immune response in the body against viruses. The selected epitopes were connected manually through bash shell script to each other using a specialized Glycine-Serine linker i.e., GGGS sequence, H-linker (HEYGAEALERAG) and PADRE Sequence (AKFVAAWTLKAAA). A four molecular adjuvant was added towards the N-terminal direction of the epitopes. These adjuvants were attached to the epitopes by one EAAAK stiff linker. Eventually, twelve vaccine models were constructed using three shortlisted epitopes. Table 4 showed the details of the vaccine constructs.
Table 4.
Twelve shortlisted vaccine models against Omicron Variant of SARs-CoV-2.
| S. No. | Position | Vaccine Model |
|---|---|---|
| 1 | Spike Protein, HBHA protein Adjuvant, and shortlisted Epitopes (320–378, 1050–1101, 1006–1043) | EAAAKMAENPNIDDLPAPLLAALGAADLALATVNDLIANLRERAEETRAETRTRVEERRARLTKFQEDLPEQFIELRDKFTTEELRKAAEGYLEAATNRYNELVERGEAALQRLRSQTAFEDASARAEGYVDQAVELTQEALGTVASQTRAVGERAAKLVGIELEAAAKAKFVAAWTLKAAAGGGSTESIVRFPNITNLCPFDEVFNATRFASVYAWNRKRISNCVADYSVLYNLAPFFTFKCYGGGGSPQSAPHGVVFLHVTYVPAQEKNFTTAPAICHDGKAHFPREGVFVSNGTHWFHEYGAEALERAGTQQLIRAAEIRASANLAATKMSECVLGQSKRVDFCGKGHEYGAEALERAGAKFVAAWTLKAAAGGGS |
| 2 | Spike Protein, HBHA conserved protein Adjuvant, and shortlisted Epitopes (320–378, 1050–1101, 1006–1043) | EAAAKMAENSNIDDIKAPLLAALGAADLALATVNELITNLRERAEETRRSRVEESRARLTKLQEDLPEQLTELREKFTAEELRKAAEGYLEAATSELVERGEAALERLRSQQSFEEVSARAEGYVDQAVELTQEALGTVASQVEGRAAKLVGIELEAAAKAKFVAAWTLKAAAGGGSTESIVRFPNITNLCPFDEVFNATRFASVYAWNRKRISNCVADYSVLYNLAPFFTFKCYGGGGSPQSAPHGVVFLHVTYVPAQEKNFTTAPAICHDGKAHFPREGVFVSNGTHWFHEYGAEALERAGTQQLIRAAEIRASANLAATKMSECVLGQSKRVDFCGKGHEYGAEALERAGAKFVAAWTLKAAAGGGS |
| 3 | Spike Protein, beta-defensin Adjuvant, and shortlisted Epitopes (320–378, 1050–1101, 1006–1043) | EAAAKGIINTLQKYYCRVRGGRCAVLSCLPKEEQIGKCSTRGRKCCRRKKEAAAKAKFVAAWTLKAAAGGGSTESIVRFPNITNLCPFDEVFNATRFASVYAWNRKRISNCVADYSVLYNLAPFFTFKCYGGGGSPQSAPHGVVFLHVTYVPAQEKNFTTAPAICHDGKAHFPREGVFVSNGTHWFHEYGAEALERAGTQQLIRAAEIRASANLAATKMSECVLGQSKRVDFCGKGHEYGAEALERAGAKFVAAWTLKAAAGGGS |
| 4 | Spike Protein, Ribosomal Adjuvant, and shortlisted Epitopes (320–378, 1050–1101, 1006–1043) | EAAAKMAKLSTDELLDAFKEMTLLELSDFVKKFEETFEVTAAAPVAVAAAGAAPAGAAVEAAEEQSEFDVILEAAGDKKIGVIKVVREIVSGLGLKEAKDLVDGAPKPLLEKVAKEAADEAKAKLEAAGATVTVKEAAAKAKFVAAWTLKAAAGGGSTESIVRFPNITNLCPFDEVFNATRFASVYAWNRKRISNCVADYSVLYNLAPFFTFKCYGGGGSPQSAPHGVVFLHVTYVPAQEKNFTTAPAICHDGKAHFPREGVFVSNGTHWFHEYGAEALERAGTQQLIRAAEIRASANLAATKMSECVLGQSKRVDFCGKGHEYGAEALERAGAKFVAAWTLKAAAGGGS |
| 5 | Spike Protein, HBHA protein Adjuvant, and shortlisted Epitopes (1006–1043, 1050–1101, 320–378) | EAAAKMAENPNIDDLPAPLLAALGAADLALATVNDLIANLRERAEETRAETRTRVEERRARLTKFQEDLPEQFIELRDKFTTEELRKAAEGYLEAATNRYNELVERGEAALQRLRSQTAFEDASARAEGYVDQAVELTQEALGTVASQTRAVGERAAKLVGIELEAAAKAKFVAAWTLKAAAGGGSTQQLIRAAEIRASANLAATKMSECVLGQSKRVDFCGKGGGGSTQQLIRAAEIRASANLAATKMSECVLGQSKRVDFCGKGGGGSPQSAPHGVVFLHVTYVPAQEKNFTTAPAICHDGKAHFPREGVFVSNGTHWFHEYGAEALERAGTESIVRFPNITNLCPFDEVFNATRFASVYAWNRKRISNCVADYSVLYNLAPFFTFKCYGHEYGAEALERAGAKFVAAWTLKAAAGGGS |
| 6 | Spike Protein, HBHA conserved protein Adjuvant, and shortlisted Epitopes (1006–1043, 1050–1101, 320–378) | EAAAKMAENSNIDDIKAPLLAALGAADLALATVNELITNLRERAEETRRSRVEESRARLTKLQEDLPEQLTELREKFTAEELRKAAEGYLEAATSELVERGEAALERLRSQQSFEEVSARAEGYVDQAVELTQEALGTVASQVEGRAAKLVGIELEAAAKAKFVAAWTLKAAAGGGSTQQLIRAAEIRASANLAATKMSECVLGQSKRVDFCGKGGGGSTQQLIRAAEIRASANLAATKMSECVLGQSKRVDFCGKGGGGSPQSAPHGVVFLHVTYVPAQEKNFTTAPAICHDGKAHFPREGVFVSNGTHWFHEYGAEALERAGTESIVRFPNITNLCPFDEVFNATRFASVYAWNRKRISNCVADYSVLYNLAPFFTFKCYGHEYGAEALERAGAKFVAAWTLKAAAGGGS |
| 7 | Spike Protein, beta-defensin Adjuvant, and shortlisted Epitopes (1006–1043, 1050–1101, 320–378) | EAAAKGIINTLQKYYCRVRGGRCAVLSCLPKEEQIGKCSTRGRKCCRRKKEAAAKAKFVAAWTLKAAAGGGSTQQLIRAAEIRASANLAATKMSECVLGQSKRVDFCGKGGGGSTQQLIRAAEIRASANLAATKMSECVLGQSKRVDFCGKGGGGSPQSAPHGVVFLHVTYVPAQEKNFTTAPAICHDGKAHFPREGVFVSNGTHWFHEYGAEALERAGTESIVRFPNITNLCPFDEVFNATRFASVYAWNRKRISNCVADYSVLYNLAPFFTFKCYGHEYGAEALERAGAKFVAAWTLKAAAGGGS |
| 8 | Spike Protein, Ribosomal Adjuvant, and shortlisted Epitopes (1006–1043, 1050–1101, 320–378) | EAAAKMAKLSTDELLDAFKEMTLLELSDFVKKFEETFEVTAAAPVAVAAAGAAPAGAAVEAAEEQSEFDVILEAAGDKKIGVIKVVREIVSGLGLKEAKDLVDGAPKPLLEKVAKEAADEAKAKLEAAGATVTVKEAAAKAKFVAAWTLKAAAGGGSTQQLIRAAEIRASANLAATKMSECVLGQSKRVDFCGKGGGGSTQQLIRAAEIRASANLAATKMSECVLGQSKRVDFCGKGGGGSPQSAPHGVVFLHVTYVPAQEKNFTTAPAICHDGKAHFPREGVFVSNGTHWFHEYGAEALERAGTESIVRFPNITNLCPFDEVFNATRFASVYAWNRKRISNCVADYSVLYNLAPFFTFKCYGHEYGAEALERAGAKFVAAWTLKAAAGGGS |
| 9 | Spike Protein, HBHA protein Adjuvant, and shortlisted Epitopes (1050–1101, 320–378, 1006–1043) | EAAAKMAENPNIDDLPAPLLAALGAADLALATVNDLIANLRERAEETRAETRTRVEERRARLTKFQEDLPEQFIELRDKFTTEELRKAAEGYLEAATNRYNELVERGEAALQRLRSQTAFEDASARAEGYVDQAVELTQEALGTVASQTRAVGERAAKLVGIELEAAAKAKFVAAWTLKAAAGGGSPQSAPHGVVFLHVTYVPAQEKNFTTAPAICHDGKAHFPREGVFVSNGTHWFGGGSTESIVRFPNITNLCPFDEVFNATRFASVYAWNRKRISNCVADYSVLYNLAPFFTFKCYGHEYGAEALERAGTQQLIRAAEIRASANLAATKMSECVLGQSKRVDFCGKGHEYGAEALERAGAKFVAAWTLKAAAGGGS |
| 10 | Spike Protein, HBHA conserved protein Adjuvant, and shortlisted Epitopes (1050–1101, 320–378, 1006–1043) | EAAAKMAENSNIDDIKAPLLAALGAADLALATVNELITNLRERAEETRRSRVEESRARLTKLQEDLPEQLTELREKFTAEELRKAAEGYLEAATSELVERGEAALERLRSQQSFEEVSARAEGYVDQAVELTQEALGTVASQVEGRAAKLVGIELEAAAKAKFVAAWTLKAAAGGGSPQSAPHGVVFLHVTYVPAQEKNFTTAPAICHDGKAHFPREGVFVSNGTHWFGGGSTESIVRFPNITNLCPFDEVFNATRFASVYAWNRKRISNCVADYSVLYNLAPFFTFKCYGHEYGAEALERAGTQQLIRAAEIRASANLAATKMSECVLGQSKRVDFCGKGHEYGAEALERAGAKFVAAWTLKAAAGGGS |
| 11 | Spike Protein, beta-defensin Adjuvant, and shortlisted Epitopes (1050–1101, 320–378, 1006–1043) | EAAAKGIINTLQKYYCRVRGGRCAVLSCLPKEEQIGKCSTRGRKCCRRKKEAAAKAKFVAAWTLKAAAGGGSPQSAPHGVVFLHVTYVPAQEKNFTTAPAICHDGKAHFPREGVFVSNGTHWFGGGSTESIVRFPNITNLCPFDEVFNATRFASVYAWNRKRISNCVADYSVLYNLAPFFTFKCYGHEYGAEALERAGTQQLIRAAEIRASANLAATKMSECVLGQSKRVDFCGKGHEYGAEALERAGAKFVAAWTLKAAAGGGS |
| 12 | Spike Protein, Ribosomal Adjuvant, and shortlisted Epitopes (1050–1101, 320–378, 1006–1043) | EAAAKMAKLSTDELLDAFKEMTLLELSDFVKKFEETFEVTAAAPVAVAAAGAAPAGAAVEAAEEQSEFDVILEAAGDKKIGVIKVVREIVSGLGLKEAKDLVDGAPKPLLEKVAKEAADEAKAKLEAAGATVTVKEAAAKAKFVAAWTLKAAAGGGSPQSAPHGVVFLHVTYVPAQEKNFTTAPAICHDGKAHFPREGVFVSNGTHWFGGGSTESIVRFPNITNLCPFDEVFNATRFASVYAWNRKRISNCVADYSVLYNLAPFFTFKCYGHEYGAEALERAGTQQLIRAAEIRASANLAATKMSECVLGQSKRVDFCGKGHEYGAEALERAGAKFVAAWTLKAAAGGGS |
The antigenicity, solubility, and allergenicity of the twelve vaccine models were evaluated further. Eight vaccine models (i.e., V2, V3, V4, V7, V8, V10, V11, and V12) were identified as highly allergic in nature using the AlgPred program with scores ranging from 0.2 to 0.3 and were eliminated. However, the antigenicity predicted by the ANTIGENpro server, and the solubility predicted through SOLpro tool of the remaining four vaccine constructs for their successful expression in E. coli vector. It resulted in the high antigenicity and solubility for remaining four vaccine constructs (V1, V5, V6, and V9) with scores ranging from 0.4 to 0.8 and were thus subjected to further investigation. The ProtParam was used to predict physicochemical features for four shortlisted vaccine designs. The estimated molecular weight of the vaccine's models was ∼41 KDa, with a pI score of ∼5, an instability index score of 28–39, and a high aliphatic score of 83–86. On the other hand, the grand average of hydropathicity was estimated to be in the range of −0.2. Nevertheless, among these four vaccines only V1 construct was identified to be stable with adjuvant HBHA. Table 5 lists the allergenicity, solubility, and antigenicity of all twelve vaccines and physico-chemical properties of four vaccine models.
Table 5.
Allergenicity, Antigenicity, Solubility, and Physicochemical Properties Analysis of twelve model vaccines.
| S. No. | Allergenicity | Antigenicity | Solubility | Amino Acids | GRAVY | Stability |
|---|---|---|---|---|---|---|
| V1 | −0.404251 | 0.4621 | 0.843915 | 397 | −0.25 | 39.96 (Stable) |
| V2 | −0.355338 | 0.4562 | 0.858297 | – | – | – |
| V3 | −0.111805 | 0.4521 | 0.782081 | – | – | – |
| V4 | 0.468878 | 0.4207 | 0.805054 | – | – | – |
| V5 | −0.440064 | 0.4789 | 0.793252 | 421 | −0.25 | 42 (Unstable) |
| V6 | −0.411146 | 0.4739 | 0.84225 | 412 | −0.22 | 46.4 (Unstable) |
| V7 | −0.191014 | 0.4764 | 0.640034 | – | – | – |
| V8 | 0.391734 | 0.4431 | 0.76977 | – | – | – |
| V9 | −0.404251 | 0.4482 | 0.832579 | 379 | −0.25 | 40 (Unstable) |
| V10 | −0.355338 | 0.4419 | 0.866211 | – | – | – |
| V11 | −0.111805 | 0.4321 | 0.768935 | – | – | – |
| V12 | 0.468878 | 0.4056 | 0.794716 | – | – | – |
3.10. Structure prediction and validation of vaccine constructs
The 3D structure of V1 vaccine construct was modeled using the SWISSMODEL tool. The template for V1 was PDB ID: 6NB3, a Spike glycoprotein from MERS-CoV complex with human neutralizing LCA60 antibody Fab fragment with 18% sequence identity, 0.25 GMQE score, and QMEANDisCo Global:0.42 ± 0.06 (Fig. 5 ). Furthermore, the stereochemical properties of the final selected vaccine construct were identified as 90.0 percent residues are in favorable region, 8.9 percent residues are in additionally allowed region, and 0.5 percent residues are in disallowed region based on the 3D structure evaluation through PROCHECK (Supplementary File 4-Fig. S1a). The ProSA program projected a Z-score of −2.52, indicating that the model is like structures determined from NMR/X-ray crystallography (Supplementary File 4-Fig. S1b). Furthermore, as shown in Supplementary File 4-Fig. S1c, the PSIPRED program was utilized to validate the 2D (secondary) structure which revealed similar number of alpha helices, beta sheets, and beta turns.
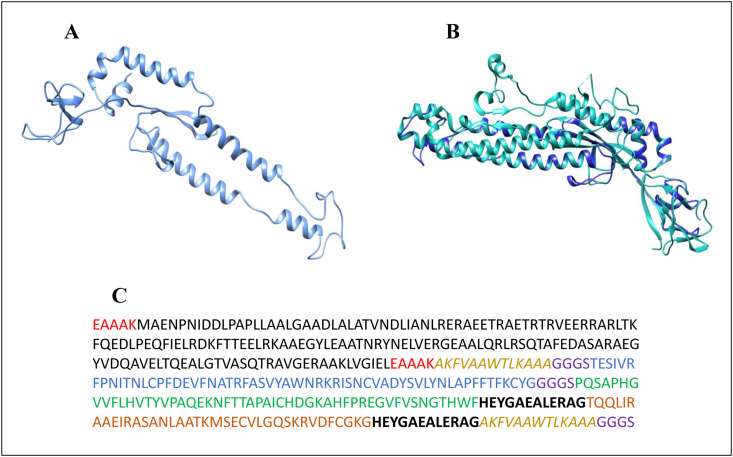
Fig. 5.
Vaccine structure modeling and Validation. (A) The 3D model of a multi-epitope vaccine was obtained by Swiss Model, (B) the vaccine sequence (Blue) superimpose on template protein (Sea Green), and (C) V1 sequence, the 397 amino acid long vaccine sequence containing adjuvant at both N and C terminal (Black) was linked with the multi-epitope sequences TESIVRFPNITNLCPFDEVFNATRFASVYAWNRKRISNCVADYSVLYNLAPFFTFKCYG (Blue), PQSAPHGVVFLHVTYVPAQEKNFTTAPAICHDGKAHFPREGVFVSNGTHWF (Green), and TQQLIRAAEIRASANLAATKMSECVLGQSKRVDFCGKG (Orange), through an EAAAK linker (red), GGGS (purple) linkers, PADRE (tan), and H-linkers (Black).
3.11. Molecular docking of vaccine construct (V1)
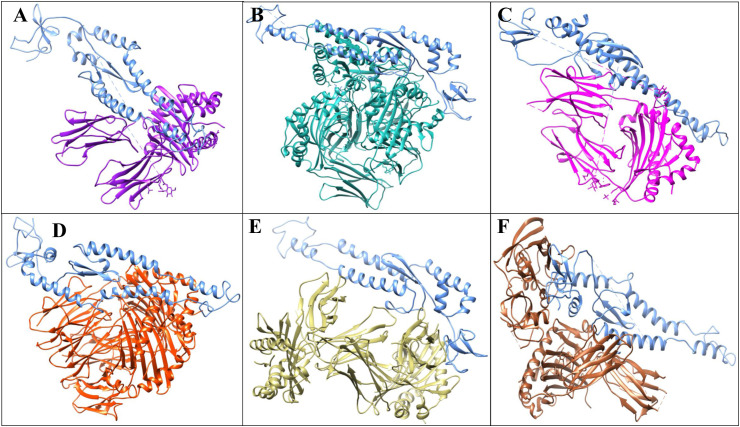
Adaptive immunity is initiated by T cells that bind to HLA molecules. The V1 vaccine was docked with the six distinct HLA alleles' protein 3C5J (HLA-DR B3*02:02), 1H15 (HLA-DR B5*01:01), 2FSE (HLA-DRB1*01:01), 2Q6W (HLA-DR B3*01:01), 2SEB (HLA-DRB1*04:01), and 1A6A (HLA-DR B1*03:01) and were then refined through FireDock tool. The docking of these HLAs and vaccine model resulted in the identification of global energies as 5.47, −15.92, −44.37, 1.21, −3.16, and −35.16 kcal/mol, respectively (Fig. 6 a). Based on these global energies, HLA-DRB1*01:01, HLA-DR B1*03:01, and HLA-DR B5*01:01 showed more potency toward V1 construct. From docking results, it can be observed that V1 may act as a potential inhibitor against Omicron variant because of high binding affinities towards HLAs.
Fig. 6a.
Docked vaccine construct with HLAs. (A) 1A6A (HLA-DR B1*03:01) (Purple) and V1 (Blue) (B) 1H15 (HLA-DR B5*01:01) (Sea Green) and V1 (Blue), (C), 3C5J (HLA-DR B3*02:02) (Magenta) and V1 (Blue), (D) 2Q6W (HLA-DR B3*01:01) (Orange) and V1 (Blue), (E) 2FSE (HLA-DRB1*01:01) (tan) and V1 (Blue), and (F) 2SEB (HLA-DRB1*04:01) (Brown) and V1 (Blue).
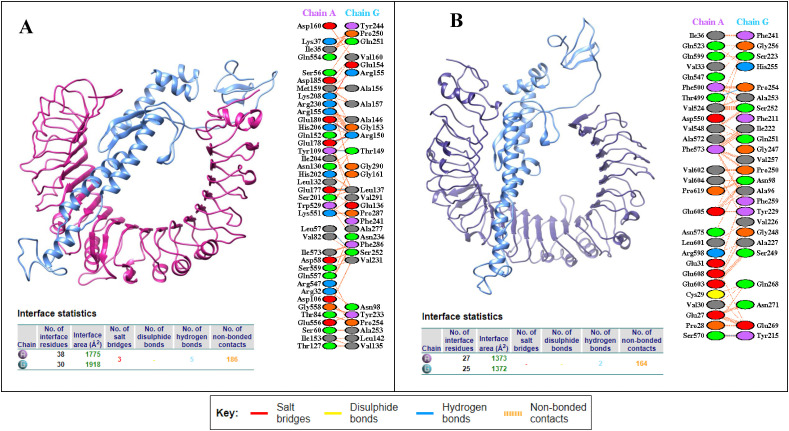
To evaluate the immunological response, the GRAMMX tool was employed to perform a docking analysis to predict the interactions between V1 and the TLR 2 (2Z7X) and TLR 4 complex (PDB 3FXI). The adjuvant HBHA protein, which serves as TLR2 and 4 agonist and produces a variety of immune responses was used to build the V1 construct. The PatchDock docking revealed binding energies of −12.12 and −1.29 kcal/mol, showing that V1 and the TLR-2/4 complex have a significant interaction (Table 6 ). However, it can be clearly observed that V1 has showed more potency towards TLR2 complex. The Protein-Protein Interactions (PPIs) of the V1 model revealed that it mediates five hydrogen bonds with TLR2, mainly involve Glu177-Leu137, Tyr109-Thr149, Glu178-Arg150, Glu180-Arg150, and Leu57-Ser252 and three salt bridges between Lys208-Arg155, Glu180-Arg150, Glu178-Arg150. Whereas two hydrogen bonds with TLR4 were observed with Thr499-Pro254, Ser570-Tyr215 as shown in Fig. 6 b.
Table 6.
Docked score of HLAs and vaccine model of omicron variant.
| Vaccine Construct | HLA alleles (PDB: ID) | SCORE | AREA | Hydrogen bond energy | Global energy | ACE |
|---|---|---|---|---|---|---|
| V1 | 1A6A | 17408 | 2658.50 | −1.93 | −35.16 | −1.83 |
| 3C5J | 17636 | 2627.70 | 0.00 | 5.47 | −0.73 | |
| 1H15 | 21712 | 3095.20 | −5.66 | −15.92 | −0.90 | |
| 2FSE | 18760 | 2695.10 | −1.31 | −44.37 | −7.34 | |
| 2Q6W | 18838 | 3802.70 | −1.12 | 1.21 | 2.17 | |
| 2SEB | 19214 | 2821.30 | −1.08 | −3.16 | 1.78 | |
| 2Z7X | 19346 | 2533.90 | −5.91 | −12.12 | 7.13 | |
| 3FXI | 24008 | 3633.10 | 0.00 | −1.29 | −0.42 |
Fig. 6b.
Docked vaccine construct with TLR2 and TLR4. (A) Docked complex of vaccine (Blue) and TLR2 (Pink), along with PPIs interactions (B) interaction occurs between the vaccine model (Blue) and TLR4 protein (Purple) along with interacting residues of vaccine represented in tables below respectively.
3.12. Molecular dynamics and immune response simulation study
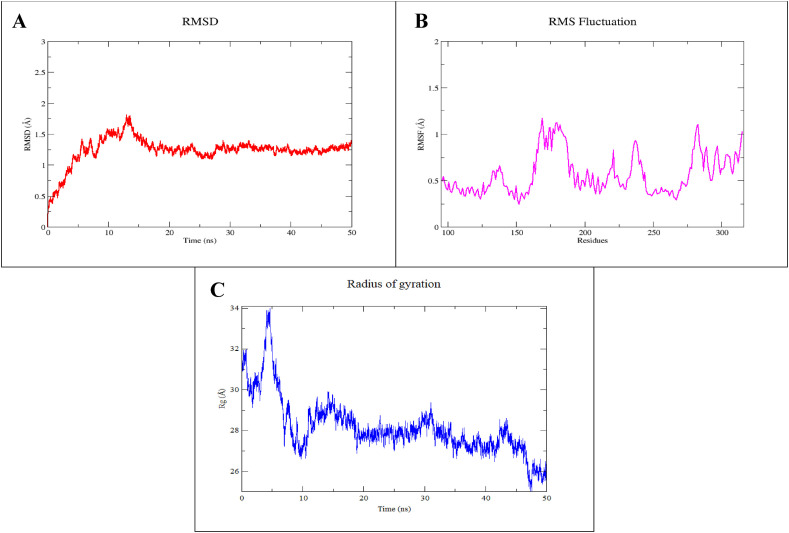
The MD simulation studies were conducted to determine the dynamics and structural stability of the designed vaccine in a time-dependent manner. The simulated trajectories were analyzed to ensure structural stability during the simulation by plotting the RMSD graph of the backbone carbon atom. As evident from Fig. 7 a, during the initial 15 ns of simulation, the system showed considerable fluctuations. However, after 15 ns the system gradually stabilized and remained stable till the end of the simulation with an average deviation of 1.23 Å that indicates the convergence of the simulated system.
Fig. 7.
Molecular dynamics simulation of V1. (A) Root mean square deviation (RMSD) of vaccine backbone (B), RMSF, and (C) Radius of gyration for vaccine model (V1) during 50 ns of MD simulation.
The predicted 3D structure of the vaccine is mostly consisted of helix and loop regions. The RMSF was plotted to further evaluate the intrinsic fluctuations of the amino acid residues. The plot showed variable fluctuations during simulation as depicted by Fig. 7b. An average RMSF was found to be 0.56 Å for the simulated system. The RMSF plot showed a few highly fluctuated regions; Ile162-Glu188, Val226-Gly247, and Val262-Gln315. All these regions were consisting of loops that are intrinsically flexible. Similarly, all the other amino acid residues of the simulated system showed significantly stable RMSF.
Furthermore, the Radius of gyration was estimated to evaluate the compactness of the vaccine structure. As evident from Fig. 7c, during the initial 5 ns of simulation, the system showed high fluctuations. However, after 10 ns the system gradually compacts, contributing to the overall stability of the simulated vaccine construct.
The deformability graph is the outcome of the Normal Mode Analysis (NMA) for the vaccine-TLRs complex's stability and mobility, according to iMODs simulation analysis. The TLR2/TLR4 and vaccine combinations were found to have eigenvalues of 1.948054e-05 and 7.526147e-06, respectively, highlighting the area of proteins with deformability depicted in terms of peaks. The variance association plot, as shown in Supplementary File 4-Fig. S2, illustrates the cumulative variance of the complex in green, while the individual variance in red and the B-factor graph resulted in a clear depiction of the docked complex. Similarly, simulation for vaccine candidate and HLAs also showed stability with an eigenvalue of 3C5J (HLA-DR B3*02:02), 1H15 (HLA-DR B5*01:01), 2FSE (HLA-DRB1*01:01), 2Q6W (HLA-DR B3*01:01), 2SEB (HLA-DRB1*04:01), and 1A6A (HLA-DR B1*03:01) were predicted as 3.54109e-05, 5.80790e-05, 4.1759e-06, 2.38221e-05, 7.07204e-06, and 1.843970e-05 respectively. It was observed that all these HLAs and TLRs especially TLR2, HLA-DR B1*03:01, HLA-DR B3*01:01, have strong affinity toward vaccine model based on their values i.e., the lower the energy the best is to deform the structure (Supplementary file 4-Figs. S3a–f).
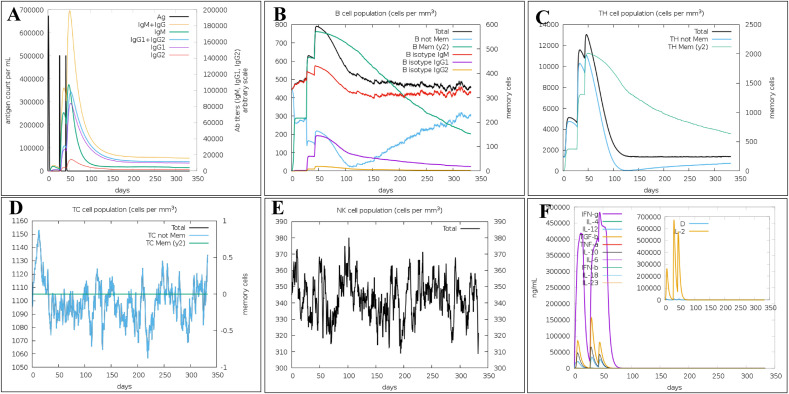
In addition, the C-Immune tool was used to predict human immune system response after vaccine injection at various time intervals. It confirmed that the immune response was consistent with the real immune reactions, such as the identification of T-cytotoxic cells, T-helper cells, and B-cells, Natural killer cells production, interleukins/interferons production, and antibody production (Fig. 7). Following the induction of vaccine injection, an increase in IgG1+IgG2, IgM, and IgG + IgM was seen, leading to a decrease in antigen concentration (Fig. 8 a and 8b). Upon vaccine construct injections, there was an increase in the production of NK cells, Th (helper), and Tc (cytotoxic) (Fig. 8c, d, and 8e). In addition, IFN-g production was increased after vaccination (Fig. 8f).
Fig. 8.
C-ImmSim presentation of an in silico immune simulation with the construct. (A) Immunoglobulin production in response to antigen injections (black vertical lines); specific subclasses are showed as colored peaks and the evolution of B-cell populations after the three injections. (B) Prediction of B cell population (C) T-helper cell populations per state after the injections. The resting state represents cells not presented with the antigen while the anergic state characterizes tolerance of the T-cells to the antigen due to repeated exposures, (D) Total production of T-cytotoxic cells (E) Natural Killer cells production levels, and (F) The main plot shows cytokine levels after the injections. The insert plot shows IL-2 level with the Simpson index; D shown by the dotted line. D is a measure of diversity. Increase in D over time indicates emergence of different epitope-specific dominant clones of T-cells. The smaller the D value, the lower the diversity.
3.13. V1 in silico cloning and codon optimization
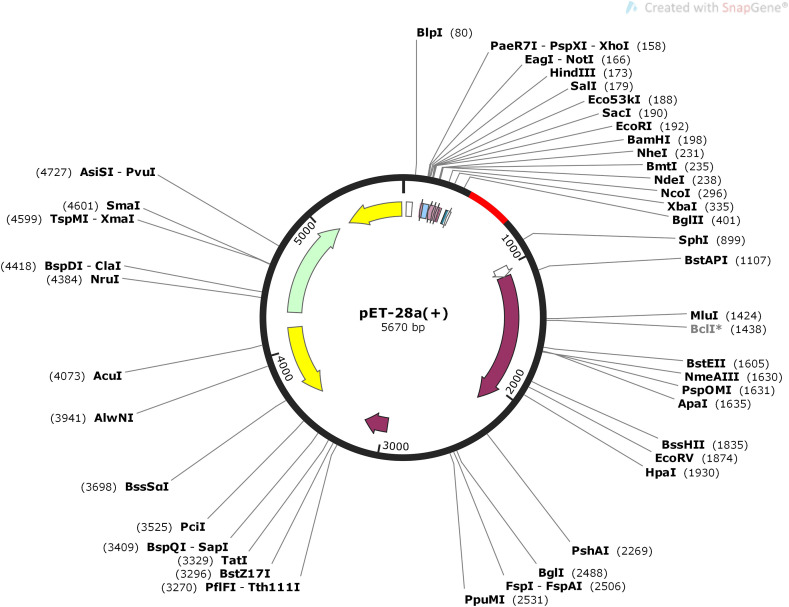
To optimize the codons and reverse translate the V1 for optimal production in E. coli (strain K12), the JCAT tool was employed. The average GC content and CAI value for V1 were estimated to be 41.9 percent and 0.61, respectively, resulting in successful vaccine construct expression. Finally, the SnapGene program was used to insert the optimized codon sequence (V1) into the pET30a (+) vector to create the recombinant plasmid (Fig. 9 ).
Fig. 9.
Codon optimization and in-silico cloning of vaccine model. In silico restriction cloning of the multi-epitope vaccine sequence into the pET30a (+) expression vector using SnapGene software, the red part represents the vaccine's gene coding, and the black circle represents the vector backbone.
4. Discussion
The COVID-19 declared as pandemic and surged to cause increase in cases and deaths due to various transmissible variants (e.g., UK (alpha/B.1.1.7), Brazil (gamma/P.1), India (delta/B.1.1.7.2) South Africa (beta/B.1.351) and now Omicron variant (B.1.1.529)) (Organization, 2021). Till the end of November 2021, the Omicron (B.1.1.529) (https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/) variant was first identified in Botswana (South Africa) along with the delta variant (B.1.1.7.2) in usually gathered genomic data that resulted in growth of case numbers and hospitalizations (Brown et al., 2021). This Variant of Concern (VOC) (https://www.cdc.gov/coronavirus/2019-ncov/variants/variant-info.html#Consequence) is changing the current pandemic trajectory by having distinct epidemiological changes in transmission rate (dominating in America, U.K (Riley et al., 2021), Scotland (Angelo et al., 2017; Sheikh et al., 2021), Israel, Sydney, and South Asia) (https://www.bbc.com/news/world-asia-53420537). A successful vaccination campaign in the start of 2021 has substantially increased immunity of population whereas the advent of the B.1.617.2 lineage of SARS-CoV-2 has put the delaying dosing tactics of immunization to a new challenge (first dose of vaccine followed by second dose given within 12 weeks) (W. Yang and Shaman, 2021). Additionally, lack of data regarding the spread of Omicron along with delta variant is currently unknown providing a huge challenge to overcome this pandemic. Nevertheless, some studies also suggested the use of multiple (three) dose regimes of vaccine to minimize such variant of concern effects and induce immunity (https://edition.cnn.com/2021/07/08/health/us-coronavirus-thursday/index.html). Due to high transmission and immune evasion of ongoing Omicron-delta (Deltacron) variant, the development of new vaccine models is urgently needed to overcome the spread of fifth wave of pandemic.
Traditional vaccine design methods use large proteins or whole organisms, which resulted in an excessive antigenic load and higher allergic responses (Chauhan et al., 2019; Sette and Fikes, 2003). Immunoinformatic techniques, on the other hand, are cost-effective and timesaving, and they may solve this problem by creating peptide-based vaccines that activate a powerful yet targeted immune response (He et al., 2018; Lu et al., 2017). As a result, the current study employed an in silico RV technique to create a multi-epitope vaccination against the Omicron spike protein (Fig. 1), which can activate immune cells (Abraham Peele et al., 2020). Multi-epitopes-based vaccine design is a new discipline that creates vaccine models that are not only protective in vivo but also effective (Cao et al., 2017; Guo et al., 2014; Zhou et al., 2009) (Jiang et al., 2017; Lennerz et al., 2014; Slingluff et al., 2013; Toledo et al., 2001).
The current work identified immunogenic MHC-I, MHC-II, and B-cell epitopes that may be used to build a multi-epitope vaccine utilizing various filters such as: (i) The epitopes must be non-toxic, antigenic, non-allergenic, and highly conserved (Table 1, Table 2), (ii) have ability to bind to MHC-I/II alleles, and should be overlapping to CTL, HTL, and B-cell epitopes (Table 3). Bazhan et al. applied the similar approach to design T-cell multi epitope vaccine model against Ebola virus that was significantly immunogenic in mice (Bazhan et al., 2019). The current work utilized four adjuvants, including beta-defensin, and L7/L12 ribosomal protein, HBHA protein, HBHA conserved sequence, as well as GGGS, PADRE sequences, HEYGAEALERAG, and EAAAK linkers, to simulate twelve distinct vaccine constructs (Table 4). These twelve vaccination models, antigenicity, solubility, allergenicity and physiochemical properties were studied to narrow down the most promiscuous vaccine construct against the Omicron variant. The V1 was identified as the most powerful vaccine construct against omicron variant as non-allergenic, most antigenic, most soluble over expressing in E. coli, and having suitable physicochemical features because of the filtering (Table 5). Similar in silico strategies were also applied by Foroutan et al. against Toxoplasma gondii to evaluate the allergenicity and physiochemical properties of their model vaccine and through laboratory validation. It was validated that this vaccine design approach was able to trigger immune response in mice (Foroutan et al., 2020). As a result, the shortlisted V1 vaccine was modeled using the SWISSMODEL program (Fig. 5), and the modeled structure was validated using PROCHECK. The Ramachandran plot revealed that 90% of residues were classified in the desired area, indicating that the vaccine's tertiary structure was verified. The protein's Z-score of −2.52 showed that it belongs to the experimentally validated structures of proteins solved by NMR and X-ray crystallographic methods, as assessed by the ProSA online site.
The spike protein of the Omicron variant must interact with Toll-Like Receptor 4 (TLR4) produced in immune cells to elicit CTB (Boehme and Compton, 2004; Carty and Bowie, 2010; Xagorari and Chlichlia, 2008). It has been reported that the CTB lost ability to trigger inflammatory response in TLR4-deficient macrophages (Vaure and Liu, 2014). It was demonstrated through ELISA-based assays that the direct binding of CTB with TLR4 inflicts the activation of NF-κB (Phongsisay et al., 2015). Additionally, the molecular docking experiments were conducted to evaluate the interaction of the modeled vaccine design with Human Leukocyte Antigen (HLA) and TLR2 and 4. The TLR2-vaccine model mainly involves five hydrogen bonds and three salt bridges among Glu177-Leu137, Tyr109-Thr149, Glu178-Arg150, Glu180-Arg150, and Leu57-Ser252, Lys208-Arg155, Glu180-Arg150, Glu178-Arg150 residues. Whereas two hydrogen bonds with TLR4 were observed with Thr499-Pro254, Ser570-Tyr215 (Fig. 6a, Fig. 6b). Several studies highlighted the importance of interaction of vaccines with TLR4 such as, Totura et al. demonstrated the susceptibility of mice to SARS-CoV infection is relatively high in TLR4 deficient mice compared to wild type (Totura et al., 2015). Similarly Hu et al. observed that upregulation in expression of TLR4 when exposed to SARS-CoV infection, suggesting the importance of TLR in immune response stimulation (Hu et al., 2012).
Importantly, the vaccine model was seen to be stable at 15 ns after a 50-ns molecular dynamics simulation (Fig. 7). The V1 model's codon optimization was reverse translated to its cDNA to enable effective expression in the E. coli pET-28a(+) expression vector. The expected GC and CAI values of V1 were 41.9 and 0.61%, respectively, indicating vaccine expression success (Fig. 9). Comparably, Foroutan et al. performed in silico codon optimization before expressing it in mice (Foroutan et al., 2020). Immune simulations of vaccine models revealed that the developed vaccine against the Omicron variant evoked a substantial immune response (Fig. 8). Correspondingly, the immune-simulation studies have been widely used for the construction of chimeric vaccine model against Klebsiella pneumoniae (Solanki et al., 2021), Mycobacterium tuberculosis (Bibi et al., 2021), Acinetobacter baumannii (Solanki and Tiwari, 2018), Ebola virus (Ullah et al., 2020) as well as against cancerous antigens (Zhang, 2018). Finally, using a reverse vaccinology technique, one vaccine construct against the Omicron version of SARS-CoV-2 was found and selected.
The proposed vaccine candidate met all of the required criteria including physiochemical characteristics, antigenicity, and non-allergenicity. As a result of this finding, we determined that our vaccine construct (V1) is safe and could be administered to humans following the completion of a successful pre-clinical and clinical trial. This V1 construct is capable of inducing an innate, cellular and humoral immune response and displayed c8onsiderable expression when back translated to cDNA in the E. coli vector pET30a(+) plasmid. Immunoinformatic has proven that the vaccine construct can stimulate a reasonable immune response. However, to ensure vaccine design efficacy in animal models and human clinical trials, in vitro and in vivo evaluation of identified V1 model is crucial. This developed vaccine candidate has the potential to help manage the next COVID-19 wave and the global pandemic crisis.
Ethical statement
This article does not contain any studies with human participants performed by any of the authors.
Funding
This study was not funded by any funding agency.
CRediT authorship contribution statement
Kanwal Khan: performed the jobs equally, Writing – original draft, jointly wrote the article. Salman Ali Khan: performed the jobs equally, Writing – original draft, jointly wrote the article. Khurshid Jalal: performed the jobs equally, Writing – original draft, jointly wrote the article. Zaheer Ul-Haq: Formal analysis, Data curation, has contributed to the analysis of the data. Reaz Uddin: Supervision, has conceived the idea and supervised the work, Writing – review & editing, has reviewed and proof-read the manuscript.
Declaration of competing interest
All Authors declare that they don't have any conflict of interest.
Acknowledgements
The authors would like to acknowledge the Higher Education Commission of Pakistan for providing the financial support under National Research Program for Universities.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.virol.2022.05.001.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- Abraham Peele K., Srihansa T., Krupanidhi S., Ayyagari V.S., Venkateswarulu T., Dynamics Design of multi-epitope vaccine candidate against SARS-CoV-2: a in-silico study. J. Biomol. Struct. Dyn. 2020;39(10):3793–3801. doi: 10.1080/07391102.2020.1770127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almofti Y.A., Abd-Elrahman K.A., Eltilib E.E. Vaccinomic approach for novel multi epitopes vaccine against severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) BMC Immunol. 2021;22(1):1–20. doi: 10.1186/s12865-021-00412-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelo M.A., Grifoni A., O'Rourke P.H., Sidney J., Paul S., Peters B., de Silva A.D., Phillips E., Mallal S., Diehl S.A. Human CD4+ T cell responses to an attenuated tetravalent dengue vaccine parallel those induced by natural infection in magnitude, HLA restriction, and antigen specificity. J. Virol. 2017;91(5) doi: 10.1128/JVI.02147-16. e02147-02116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araújo C.L., Alves J., Nogueira W., Pereira L.C., Gomide A.C., Ramos R., Azevedo V., Silva A., Folador A. Prediction of new vaccine targets in the core genome of Corynebacterium pseudotuberculosis through omics approaches and reverse vaccinology. Gene. 2019;702:36–45. doi: 10.1016/j.gene.2019.03.049. [DOI] [PubMed] [Google Scholar]
- Bahrami A.A., Payandeh Z., Khalili S., Zakeri A., Bandehpour M. Immunoinformatics: in Silico approaches and computational design of a multi-epitope, immunogenic protein. Int. Rev. Immunol. 2019;38(6):307–322. doi: 10.1080/08830185.2019.1657426. [DOI] [PubMed] [Google Scholar]
- Bazhan S.I., Antonets D.V., Karpenko L.I., Oreshkova S.F., Kaplina O.N., Starostina E.V., Dudko S.G., Fedotova S.A., Ilyichev A.A. In silico designed ebola virus T-cell multi-epitope DNA vaccine constructions are immunogenic in mice. Vaccines. 2019;7(2):34. doi: 10.3390/vaccines7020034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya M., Malick R.C., Mondal N., Patra P., Pal B.B., Patra B.C., Das B.K. Computational characterization of epitopic region within the outer membrane protein candidate in Flavobacterium columnare for vaccine development. J. Biomol. Struct. Dyn. 2019;38(2):450–459. doi: 10.1080/07391102.2019.1580222. [DOI] [PubMed] [Google Scholar]
- Bibi S., Ullah I., Zhu B., Adnan M., Liaqat R., Kong W.-B., Niu S. In silico analysis of epitope-based vaccine candidate against tuberculosis using reverse vaccinology. Sci. Rep. 2021;11(1):1–16. doi: 10.1038/s41598-020-80899-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehme K.W., Compton T. Innate sensing of viruses by toll-like receptors. J. Virol. 2004;78(15):7867–7873. doi: 10.1128/JVI.78.15.7867-7873.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown K.A., Gubbay J., Buchan S.A., Daneman N., Mishra S., Patel S., Day T. Canada. medRxiv; 2021. Inflection in Prevalence of SARS-CoV-2 Infections Missing the N501Y Mutation as a Marker of Rapid Delta (B. 1.617. 2) Lineage Expansion in Ontario. [Google Scholar]
- Burley S.K., Bhikadiya C., Bi C., Bittrich S., Chen L., Crichlow G.V., Christie C.H., Dalenberg K., Di Costanzo L., Duarte J.M. RCSB Protein Data Bank: powerful new tools for exploring 3D structures of biological macromolecules for basic and applied research and education in fundamental biology, biomedicine, biotechnology, bioengineering and energy sciences. Nucleic Acids Res. 2021;49(D1):D437–D451. doi: 10.1093/nar/gkaa1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Li D., Fu Y., Bai Q., Chen Y., Bai X., Jing Z., Sun P., Bao H., Li P. Rational design and efficacy of a multi-epitope recombinant protein vaccine against foot-and-mouth disease virus serotype A in pigs. Antivir. Res. 2017;140:133–141. doi: 10.1016/j.antiviral.2017.01.023. [DOI] [PubMed] [Google Scholar]
- Carty M., Bowie A.G. Recent insights into the role of Toll‐like receptors in viral infection. Clin. Exp. Immunol. 2010;161(3):397–406. doi: 10.1111/j.1365-2249.2010.04196.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan V., Rungta T., Goyal K., Singh M.P. Designing a multi-epitope based vaccine to combat Kaposi Sarcoma utilizing immunoinformatics approach. Sci. Rep. 2019;9(1):1–15. doi: 10.1038/s41598-019-39299-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou P. Analysis of the accuracy and implications of simple method for predicting the secondary structure of globular proteins. J. Adv. Enzym. 1978;47:45–148. [Google Scholar]
- Dhanda S.K., Mahajan S., Paul S., Yan Z., Kim H., Jespersen M.C., Jurtz V., Andreatta M., Greenbaum J.A., Marcatili P. IEDB-AR: immune epitope database—analysis resource in 2019. Nucleic Acids Res. 2019;47(W1):W502–W506. doi: 10.1093/nar/gkz452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doytchinova I.A., Flower D.R. VaxiJen: a server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinf. 2007;8(1):1–7. doi: 10.1186/1471-2105-8-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doytchinova I.A., Guan P., Flower D.R. EpiJen: a server for multistep T cell epitope prediction. BMC Bioinf. 2006;7(1):1–11. doi: 10.1186/1471-2105-7-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El‐Manzalawy Y., Dobbs D., Honavar V. Predicting linear B‐cell epitopes using string kernels. J. Mol. Recogn. 2008;21(4):243–255. doi: 10.1002/jmr.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esmailnia E., Amani J., Gargari S.L.M. Identification of novel vaccine candidate against Salmonella enterica serovar Typhi by reverse vaccinology method and evaluation of its immunization. Genomics. 2020;112(5):3374–3381. doi: 10.1016/j.ygeno.2020.06.022. [DOI] [PubMed] [Google Scholar]
- Foroutan M., Ghaffarifar F., Sharifi Z., Dalimi A.J. C.i. Vaccination with a novel multi-epitope ROP8 DNA vaccine against acute Toxoplasma gondii infection induces strong B and T cell responses in mice. Comp. Immunol. Microbiol. Infect. Dis. 2020;69 doi: 10.1016/j.cimid.2020.101413. microbiology. [DOI] [PubMed] [Google Scholar]
- Garg V.K., Avashthi H., Tiwari A., Jain P.A., Ramkete P.W., Kayastha A.M., Singh V.K. MFPPI–multi FASTA ProtParam interface. Bioinformation. 2016;12(2):74. doi: 10.6026/97320630012074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaffari-Nazari H., Tavakkol-Afshari J., Jaafari M.R., Tahaghoghi-Hajghorbani S., Masoumi E., Jalali S.A. Improving multi-epitope long peptide vaccine potency by using a strategy that enhances CD4+ T help in BALB/c mice. PLoS One. 2015;10(11) doi: 10.1371/journal.pone.0142563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grote A., Hiller K., Scheer M., Münch R., Nörtemann B., Hempel D.C., Jahn D. JCat: a novel tool to adapt codon usage of a target gene to its potential expression host. Nucleic Acids Res. 2005;33(Suppl. l_2):W526–W531. doi: 10.1093/nar/gki376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L., Yin R., Liu K., Lv X., Li Y., Duan X., Chu Y., Xi T., Xing Y. Immunological features and efficacy of a multi-epitope vaccine CTB-UE against H. pylori in BALB/c mice model. Appl. Microbiol. Biotechnol. 2014;98(8):3495–3507. doi: 10.1007/s00253-013-5408-6. [DOI] [PubMed] [Google Scholar]
- Gupta S., Kapoor P., Chaudhary K., Gautam A., Kumar R., Consortium O.S.D.D., Raghava G.P. In silico approach for predicting toxicity of peptides and proteins. PLoS One. 2013;8(9) doi: 10.1371/journal.pone.0073957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajialibeigi A., Amani J., Gargari S.L.M. Identification and evaluation of novel vaccine candidates against Shigella flexneri through reverse vaccinology approach. Appl. Microbiol. Biotechnol. 2021;105(3):1159–1173. doi: 10.1007/s00253-020-11054-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison A.G., Lin T., Wang P. Mechanisms of SARS-CoV-2 transmission and pathogenesis. Trends Immunol. 2020;41(12):1100–1115. doi: 10.1016/j.it.2020.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey W.T., Carabelli A.M., Jackson B., Gupta R.K., Thomson E.C., Harrison E.M., Ludden C., Reeve R., Rambaut A., Peacock S.J. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021;19(7):409–424. doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan M.A., Khan M.A., Sharmin T., Mazumder M.H.H., Chowdhury A.S. Identification of putative drug targets in Vancomycin-resistant Staphylococcus aureus (VRSA) using computer aided protein data analysis. Gene. 2016;575(1):132–143. doi: 10.1016/j.gene.2015.08.044. [DOI] [PubMed] [Google Scholar]
- He R., Yang X., Liu C., Chen X., Wang L., Xiao M., Ye J., Wu Y., Ye L. Efficient control of chronic LCMV infection by a CD4 T cell epitope-based heterologous prime-boost vaccination in a murine model. Cell. Mol. Immunol. 2018;15(9):815–826. doi: 10.1038/cmi.2017.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess E., Jinnah H., Kozak C., Wilson M. Spontaneous locomotor hyperactivity in a mouse mutant with a deletion including the Snap gene on chromosome 2. J. Neurosci. 1992;12(7):2865–2874. doi: 10.1523/JNEUROSCI.12-07-02865.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W., Yen Y.-T., Singh S., Kao C.-L., Wu-Hsieh B.A. SARS-CoV regulates immune function-related gene expression in human monocytic cells. Viral Immunol. 2012;25(4):277–288. doi: 10.1089/vim.2011.0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalal K., Khan K., Ahmad D., Hayat A., Basharat Z., Abbas M.N., Alghamdi S., Almehmadi M., Sahibzada M.U.K. Pan-genome reverse vaccinology approach for the design of multi-epitope vaccine construct against Escherichia albertii. Int. J. Mol. Sci. 2021;22(23) doi: 10.3390/ijms222312814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang P., Cai Y., Chen J., Ye X., Mao S., Zhu S., Xue X., Chen S., Zhang L. Evaluation of tandem Chlamydia trachomatis MOMP multi-epitopes vaccine in BALB/c mice model. Vaccine. 2017;35(23):3096–3103. doi: 10.1016/j.vaccine.2017.04.031. [DOI] [PubMed] [Google Scholar]
- Kaba S.A., Karch C.P., Seth L., Ferlez K.M., Storme C.K., Pesavento D.M., Laughlin P.Y., Bergmann-Leitner E.S., Burkhard P., Lanar D.E. Self-assembling protein nanoparticles with built-in flagellin domains increases protective efficacy of a Plasmodium falciparum based vaccine. Vaccine. 2018;36(6):906–914. doi: 10.1016/j.vaccine.2017.12.001. [DOI] [PubMed] [Google Scholar]
- Karplus P., Schulz G. Prediction of chain flexibility in proteins. Naturwissenschaften. 1985;72(4):212–213. [Google Scholar]
- Khalid K., Irum S., Ullah S.R., Andleeb S. In-silico vaccine design based on a novel vaccine candidate against infections caused by acinetobacter baumannii. Int. J. Pept. Res. Therapeut. 2022;28(1):1–17. doi: 10.1007/s10989-021-10316-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Ponomarenko J., Zhu Z., Tamang D., Wang P., Greenbaum J., Lundegaard C., Sette A., Lund O., Bourne P.E. Immune epitope database analysis resource. Nucleic Acids Res. 2012;40(W1):W525–W530. doi: 10.1093/nar/gks438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A., Parashar R., Kumar S., Faiq M.A., Kumari C., Kulandhasamy M., Narayan R.K., Jha R.K., Singh H.N., Prasoon P. Emerging SARS‐CoV‐2 variants can potentially break set epidemiological barriers in COVID‐19. J. Med. Virol. 2021 doi: 10.1002/jmv.27467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski R.A. PDBsum: summaries and analyses of PDB structures. Nucleic Acids Res. 2001;29(1):221–222. doi: 10.1093/nar/29.1.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennerz V., Gross S., Gallerani E., Sessa C., Mach N., Boehm S., Hess D., Von Boehmer L., Knuth A., Ochsenbein A.F. Immunologic response to the survivin-derived multi-epitope vaccine EMD640744 in patients with advanced solid tumors. Cancer Immunol. Immunother. 2014;63(4):381–394. doi: 10.1007/s00262-013-1516-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Lai S., Gao G.F., Shi W. The emergence, genomic diversity and global spread of SARS-CoV-2. Nature. 2021:1–11. doi: 10.1038/s41586-021-04188-6. [DOI] [PubMed] [Google Scholar]
- Lin Z., van Gunsteren W.F. Refinement of the application of the GROMOS 54A7 force field to β‐peptides. J. Comput. Chem. 2013;34(32):2796–2805. doi: 10.1002/jcc.23459. [DOI] [PubMed] [Google Scholar]
- Lu C., Meng S., Jin Y., Zhang W., Li Z., Wang F., Wang‐Johanning F., Wei Y., Liu H., Tu H. A novel multi‐epitope vaccine from MMSA‐1 and DKK 1 for multiple myeloma immunotherapy. Br. J. Haematol. 2017;178(3):413–426. doi: 10.1111/bjh.14686. [DOI] [PubMed] [Google Scholar]
- Morris G.M., Lim-Wilby M. J. Mol. Model. Springer; 2008. Molecular docking; pp. 365–382. [DOI] [PubMed] [Google Scholar]
- Moxon R., Reche P.A., Rappuoli R. Reverse vaccinology. Front. Immunol. 2019;10:2776. doi: 10.3389/fimmu.2019.02776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiura H., Ito K., Anzai A., Kobayashi T., Piantham C., Rodríguez-Morales A.J. Relative reproduction number of SARS-CoV-2 omicron (B. 1.1. 529) compared with delta variant in South Africa. J. Clin. Med. 2022;11(1):30. doi: 10.3390/jcm11010030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Organization W.H. 2021. Interim Recommendations for Use of the Pfizer–BioNTech COVID-19 Vaccine, BNT162b2, under Emergency Use Listing: Interim Guidance. first issued 8 January 2021, updated 15 June 2021. Retrieved from. [Google Scholar]
- Parker J., Guo D., Hodges R. New hydrophilicity scale derived from high-performance liquid chromatography peptide retention data: correlation of predicted surface residues with antigenicity and X-ray-derived accessible sites. Biochemistry. 1986;25(19):5425–5432. doi: 10.1021/bi00367a013. [DOI] [PubMed] [Google Scholar]
- Petersen E., Ntoumi F., Hui D.S., Abubakar A., Kramer L.D., Obiero C., Tambyah P.A., Blumberg L., Yapi R., Al-Abri S. Emergence of new SARS-CoV-2 Variant of Concern Omicron (B. 1.1. 529)-highlights Africa's research capabilities, but exposes major knowledge gaps, inequities of vaccine distribution, inadequacies in global COVID-19 response and control efforts. Int. J. Infect. Dis. 2022;114:268–272. doi: 10.1016/j.ijid.2021.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Phongsisay V., Iizasa E.i., Hara H., Yoshida H. Evidence for TLR4 and FcRγ–CARD9 activation by cholera toxin B subunit and its direct bindings to TREM2 and LMIR5 receptors. Mol. Immunol. 2015;66(2):463–471. doi: 10.1016/j.molimm.2015.05.008. [DOI] [PubMed] [Google Scholar]
- Ponomarenko J., Bui H.-H., Li W., Fusseder N., Bourne P.E., Sette A., Peters B. ElliPro: a new structure-based tool for the prediction of antibody epitopes. BMC Bioinf. 2008;9(1):1–8. doi: 10.1186/1471-2105-9-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon L.L., Peiris M. Emergence of a novel human coronavirus threatening human health. Nat. Med. 2020;26(3):317–319. doi: 10.1038/s41591-020-0796-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poudel S., Ishak A., Perez-Fernandez J., Garcia E., León-Figueroa D.A., Romaní L., Bonilla-Aldana D.K., Rodriguez-Morales A.J. Highly mutated SARS-CoV-2 Omicron variant sparks significant concern among global experts–What is known so far? Trav. Med. Infect. Dis. 2022;45 doi: 10.1016/j.tmaid.2021.102234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman N., Ajmal A., Ali F., Rastrelli L. Core proteome mediated therapeutic target mining and multi-epitope vaccine design for Helicobacter pylori. Genomics. 2020;112(5):3473–3483. doi: 10.1016/j.ygeno.2020.06.026. [DOI] [PubMed] [Google Scholar]
- Rahman N., Ali F., Basharat Z., Shehroz M., Khan M.K., Jeandet P., Nepovimova E., Kuca K., Khan H. Vaccine design from the ensemble of surface glycoprotein epitopes of SARS-CoV-2: an immunoinformatics approach. Vaccines. 2020;8(3):423. doi: 10.3390/vaccines8030423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed S.G., Orr M.T., Fox C.B. Key roles of adjuvants in modern vaccines. Nat. Med. 2013;19(12):1597–1608. doi: 10.1038/nm.3409. [DOI] [PubMed] [Google Scholar]
- Riley S., Walters C.E., Wang H., Eales O., Haw D., Ainslie K.E., Atchinson C., Fronterre C., Diggle P.J., Page A.J. medRxiv; 2021. REACT-1 Round 12 Report: Resurgence of SARS-CoV-2 Infections in England Associated with Increased Frequency of the Delta Variant. [Google Scholar]
- Saha S., Raghava G.P.S. Prediction of continuous B‐cell epitopes in an antigen using recurrent neural network. Proteins. 2006;65(1):40–48. doi: 10.1002/prot.21078. [DOI] [PubMed] [Google Scholar]
- Schneidman-Duhovny D., Inbar Y., Nussinov R., Wolfson H.J. PatchDock and SymmDock: servers for rigid and symmetric docking. Nucleic Acids Res. 2005;33(Suppl. l_2):W363–W367. doi: 10.1093/nar/gki481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sette A., Fikes J. Epitope-based vaccines: an update on epitope identification, vaccine design and delivery. Curr. Opin. 2003;15(4):461–470. doi: 10.1016/s0952-7915(03)00083-9. [DOI] [PubMed] [Google Scholar]
- Sheikh A., McMenamin J., Taylor B., Robertson C. SARS-CoV-2 Delta VOC in Scotland: demographics, risk of hospital admission, and vaccine effectiveness. Lancet. 2021;397(10293):P2461–P2462. doi: 10.1016/S0140-6736(21)01358-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slingluff C.L., Lee S., Zhao F., Chianese-Bullock K.A., Olson W.C., Butterfield L.H., Whiteside T.L., Leming P.D., Kirkwood J.M. A randomized phase II trial of multiepitope vaccination with melanoma peptides for cytotoxic T cells and helper T cells for patients with metastatic melanoma (E1602) Clin. Cancer Res. 2013;19(15):4228–4238. doi: 10.1158/1078-0432.CCR-13-0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solanki V., Sharma S., Tiwari V. Subtractive proteomics and reverse vaccinology strategies for designing a multiepitope vaccine targeting membrane proteins of Klebsiella pneumoniae. Int. J. Pept. Res. Therapeut. 2021;27(2):1177–1195. [Google Scholar]
- Solanki V., Tiwari V. Subtractive proteomics to identify novel drug targets and reverse vaccinology for the development of chimeric vaccine against Acinetobacter baumannii. Sci. Rep. 2018;8(1):1–19. doi: 10.1038/s41598-018-26689-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao K., Tzou P.L., Nouhin J., Gupta R.K., de Oliveira T., Kosakovsky Pond S.L., Fera D., Shafer R.W. The biological and clinical significance of emerging SARS-CoV-2 variants. Nat. Rev. Genet. 2021;22(12):757–773. doi: 10.1038/s41576-021-00408-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur V., Bhola S., Thakur P., Patel S.K.S., Kulshrestha S., Ratho R.K., Kumar P. Waves and variants of SARS-CoV-2: understanding the causes and effect of the COVID-19 catastrophe. Infection. 2021:1–16. doi: 10.1007/s15010-021-01734-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomsen M., Lundegaard C., Buus S., Lund O., Nielsen M. MHCcluster, a method for functional clustering of MHC molecules. Immunogenetics. 2013;65(9):655–665. doi: 10.1007/s00251-013-0714-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo H., Baly A., Castro O., Resik S., Laferté J., Rolo F., Navea L., Lobaina L., Cruz O., Miguez J. A phase I clinical trial of a multi-epitope polypeptide TAB9 combined with Montanide ISA 720 adjuvant in non-HIV-1 infected human volunteers. Vaccine. 2001;19(30):4328–4336. doi: 10.1016/s0264-410x(01)00111-6. [DOI] [PubMed] [Google Scholar]
- Totura A.L., Whitmore A., Agnihothram S., Schäfer A., Katze M.G., Heise M.T., Baric R.S. Toll-like receptor 3 signaling via TRIF contributes to a protective innate immune response to severe acute respiratory syndrome coronavirus infection. mBio. 2015;6(3) doi: 10.1128/mBio.00638-15. e00638-00615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tovchigrechko A., Vakser I.A. GRAMM-X public web server for protein–protein docking. Nucleic Acids Res. 2006;34(Suppl. l_2):W310–W314. doi: 10.1093/nar/gkl206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullah M.A., Sarkar B., Islam S.S. Exploiting the reverse vaccinology approach to design novel subunit vaccines against Ebola virus. Immunobiology. 2020;225(3) doi: 10.1016/j.imbio.2020.151949. [DOI] [PubMed] [Google Scholar]
- Van Der Spoel D., Lindahl E., Hess B., Groenhof G., Mark A.E., Berendsen H.J. GROMACS: fast, flexible, and free. J. Comput. Chem. 2005;26(16):1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- Vaure C., Liu Y. A comparative review of toll-like receptor 4 expression and functionality in different animal species. Front. Immunol. 2014;5:316. doi: 10.3389/fimmu.2014.00316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Liu J., Guo S., Liu L., Yuan Q., Guo L., Pan S. Identification of Vibrio parahaemolyticus and Vibrio Spp. specific outer membrane proteins by reverse vaccinology and surface proteome. Front. Microbiol. 2020;11:3529. doi: 10.3389/fmicb.2020.625315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xagorari A., Chlichlia K. Toll-like receptors and viruses: induction of innate antiviral immune responses. Open Microbiol. J. 2008;2:49. doi: 10.2174/1874285800802010049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W., Shaman J. medRxiv; 2021. COVID-19 Pandemic Dynamics in India and Impact of the SARS-CoV-2 Delta (B. 1.617. 2) Variant. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Sun W., Guo J., Zhao G., Sun S., Yu H., Guo Y., Li J., Jin X., Du L. In silico design of a DNA-based HIV-1 multi-epitope vaccine for Chinese populations. Hum. Vaccines Immunother. 2015;11(3):795–805. doi: 10.1080/21645515.2015.1012017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yukeswaran L., Shreeranjana S., Subhashini T. Immunoinformatics aided multi-epitope based vaccine design against crimean-Congo virus. AIJR Abstracts. 2021;43 [Google Scholar]
- Zhang L. Multi-epitope vaccines: a promising strategy against tumors and viral infections. Cell. Mol. Immunol. 2018;15(2):182–184. doi: 10.1038/cmi.2017.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W.-Y., Shi Y., Wu C., Zhang W.-J., Mao X.-H., Guo G., Li H.-X., Zou Q.-M. Therapeutic efficacy of a multi-epitope vaccine against Helicobacter pylori infection in BALB/c mice model. Vaccine. 2009;27(36):5013–5019. doi: 10.1016/j.vaccine.2009.05.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.