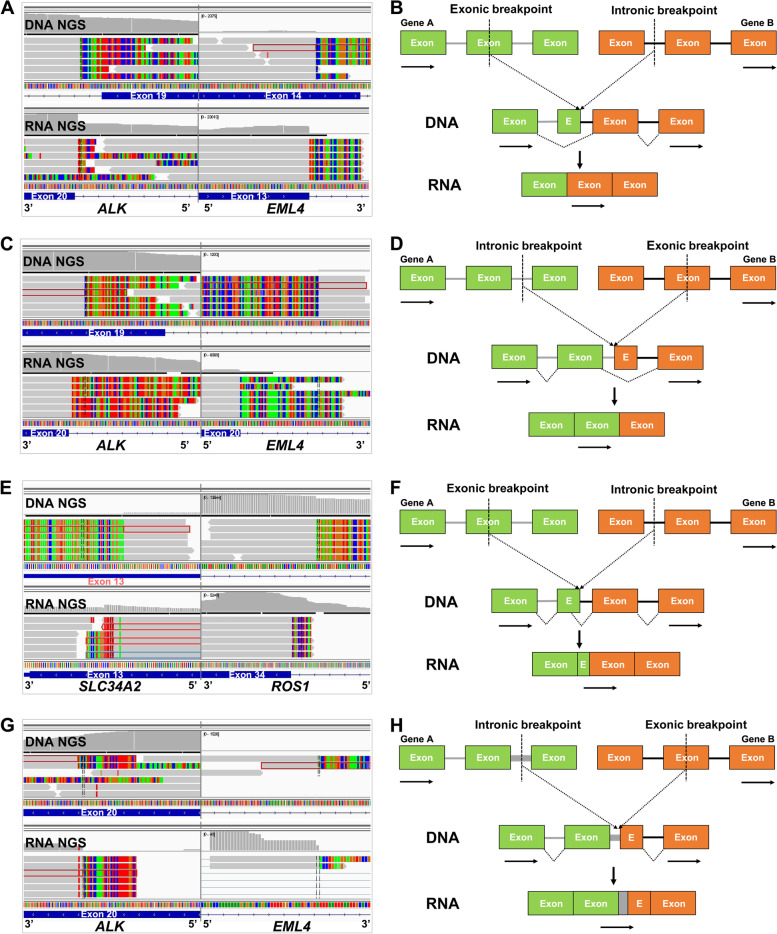
Fig. 3.
Examples and schematic diagrams of exonic-breakpoint fusions generating functional transcripts. A In P12, DNA NGS detected an exonic-breakpoint ALK fusion involving EML4 exon 14 and ALK intron 19, whereas RNA NGS revealed an ALK fusion involving EML4 exon 13 and ALK exon 20. B Schematic diagram showing that the breakpoint at the transcript level differs from that predicted by the genomic breakpoint in the “exon-intron” fusion due to exon skipping. C In P23, DNA NGS showed an exonic-breakpoint ALK fusion involving EML4 intron 20 and ALK exon 19, whereas RNA NGS detected an ALK fusion involving EML4 exon 20 and ALK exon 20. D Schematic diagram showing that the breakpoint at the transcript level differs from that predicted by the genomic breakpoint in the “intron-exon” fusion due to exon skipping. E In P37, DNA NGS showed an exonic-breakpoint ROS1 fusion involving SLC34A2 exon 13 and ROS1 intron 33, and RNA NGS revealed a ROS1 fusion involving SLC34A2 exon 13 and ROS1 exon 34. F Schematic diagram showing that the breakpoint at the transcript level differs from that predicted by the genomic breakpoint due to alternative splice site selection. G In P38, DNA NGS detected an exonic-breakpoint ALK fusion involving EML4 intron 6 and ALK exon 20, whereas RNA NGS revealed an ALK fusion involving EML4 intron 6 and ALK exon 20. H Schematic diagram showing that the breakpoint at the transcript level differs from that predicted by the genomic breakpoint due to intron retention. The gray bars indicate sequencing reads that match the reference genome, and multicolored bars indicate mismatched reads (the corresponding partners). The rectangles indicate exons (E, exon), and the solid lines indicate introns. The arrows indicate the direction of transcription