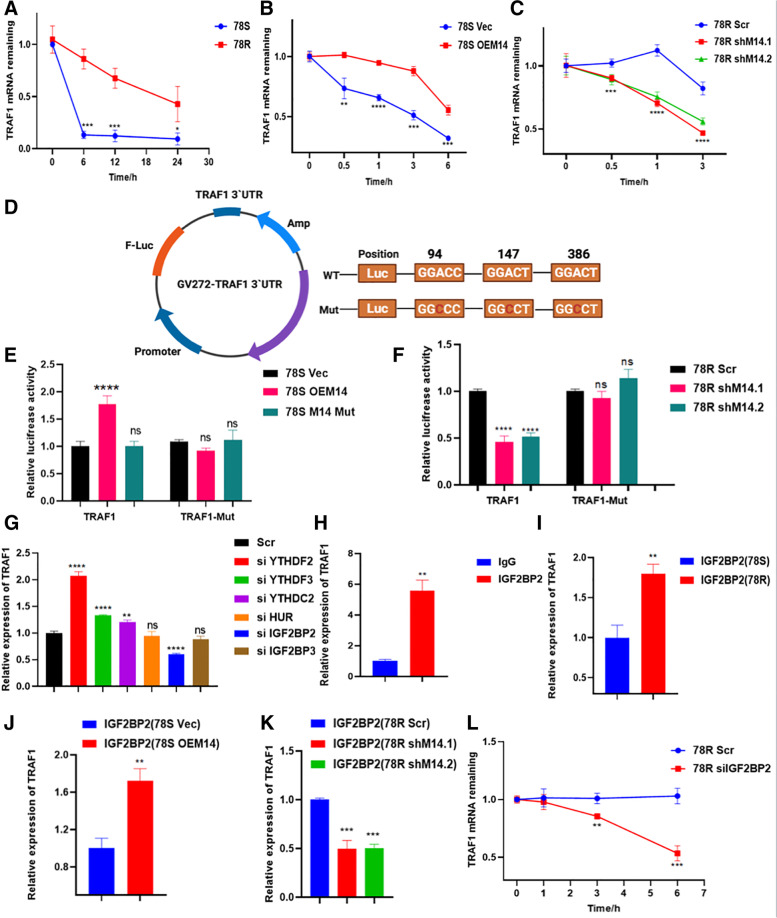
Fig. 6.
METTL14 regulates mRNA stability of TRAF1. A The mRNA half-life (t1/2) of TRAF1 transcripts in 78S and 78R cell lines. B The mRNA half-life (t1/2) of TRAF1 transcripts in 78S cells with (OEM14) or without (Vec) METTL14 overexpression. C The mRNA half-life (t1/2) of TRAF1 transcripts in 78R cells with (shM14.1 or shM14.2) or without (Scr) METTL14 depletion. D TRAF1 3′-UTR plasmid contain wild-type or mutant seed sequences. E Relative luciferase activity of TRAF1 3′-UTR constructs containing wild-type or mutant seed sequences after co-transfection with vector (Vec), METTL14 overexpression (OEM14) or mutant METTL14 overexpression (M14 Mut) into 78S cells. F Relative luciferase activity of TRAF1 3′-UTR constructs containing wild-type or mutant seed sequences after co-transfection with scramble (Scr) or shMETTL14(shM14) into 78R cells. Firefly luciferase activity was measured and normalized to Renilla luciferase activity. G qPCR analysis of the mRNA levels of TRAF1 after knock-down of readers. IGF2BP2 was screened out to positively influence the expression of TRAF1. IGF2BP2 was screened out to positively influence the expression of TRAF1. H and I RIP assay for the enrichment of TRAF1 in 78S and 78R incubated with IGF2BP2 antibody. TRAF1 is highly enriched in 78R cells compared to 78S cells. J and K RIP assay for the enrichment of TRAF1 in 78S cells with METTL14 overexpression (J) and in 78R cells with METTL14 depletion(K) incubated with IGF2BP2 antibody. L The decay rate of mRNA and qPCR analysis of TRAF1 at the indicated times after actinomycin D (5 μg/ml) treatment in 78R cells with IGF2BP2 knockdown