Abstract
T cells are thought to be an important correlates of protection against SARS‐CoV2 infection. However, the composition of T cell subsets in convalescent individuals of SARS‐CoV2 infection has not been well studied. The authors determined the lymphocyte absolute counts, the frequency of memory T cell subsets, and the plasma levels of common γ−chain in 7 groups of COVID‐19 individuals, based on days since RT‐PCR confirmation of SARS‐CoV‐2 infection. The data show that both absolute counts and frequencies of lymphocytes as well as, the frequencies of CD4+ central and effector memory cells increased, and the frequencies of CD4+ naïve T cells, transitional memory, stem cell memory T cells, and regulatory cells decreased from Days 15–30 to Days 61–90 and plateaued thereafter. In addition, the frequencies of CD8+ central memory, effector, and terminal effector memory T cells increased, and the frequencies of CD8+ naïve cells, transitional memory, and stem cell memory T cells decreased from Days 15–30 to Days 61–90 and plateaued thereafter. The plasma levels of IL‐2, IL‐7, IL‐15, and IL‐21—common γc cytokines started decreasing from Days 15–30 till Days 151–180. Severe COVID‐19 patients exhibit decreased levels of lymphocyte counts and frequencies, higher frequencies of naïve cells, regulatory T cells, lower frequencies of central memory, effector memory, and stem cell memory, and elevated plasma levels of IL‐2, IL‐7, IL‐15, and IL‐21. Finally, there was a significant correlation between memory T cell subsets and common γc cytokines. Thus, the study provides evidence of alterations in lymphocyte counts, memory T cell subset frequencies, and common γ−chain cytokines in convalescent COVID‐19 individuals.
Keywords: Memory T cell subsets; CD4+ T cell subsets; CD8+ T cell subsets; COVID‐19, acute and convalescent COVID‐19
Graphical Abstract
Our study provides evidence of alterations in lymphocyte counts, memory T cell subset frequencies and common γ‐chain cytokines in convalescent COVID‐19 individuals.
1. INTRODUCTION
Severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) is the cause for the Coronavirus disease 2019 (COVID‐19) that affects individuals globally. For the most part COVID‐19 infections are mild with recovery within 2–3 weeks. 1 However, a significant number of people progress to severe disease due to exaggerated immune response and pathology. 2 , 3 The adaptive immune system, mainly, T cells play an important role in the clearance of viral infections. 4
Mild and severe incidents of COVID‐19 are linked with a dramatically decreased total number of lymphocytes. 5 CD4+ T cell and CD8+ T cell responses might be crucial in SARS‐CoV‐2, for control and protection against primary SARS‐CoV‐2 infection. 6 Immune memory, after primary infection or immunization, is the basis of defensive immunity following a later infection. 7
The common‐γ‐chain cytokines plays a key role in health and disease. 8 Common cytokine receptor γ‐chain family (γc cytokines) are linked with the development of memory T cell generation. 9 , 10 Studies showed that, T cell development and maintenance and induction of T cell responses requires IL‐2, IL‐7, IL‐15, and IL‐21. 11 Lucas et al. exhibited that IL‐2, IL‐7, and IL‐15 were enhanced in COVID‐19 and associated with disease severity 12 and could stimulate IFN‐γ secretion by an antigen‐independent manner. 13 However, the impact of COVID‐19 on common γc cytokines IL‐2, IL‐7, IL‐15, and IL‐21‐ levels have not been well studied in COVID‐19.
Examining the intricacies of immune memory to SARS‐CoV‐2 is an important prerequisite to understand the duration of defensive immunity to COVID‐19 formed by primary SARS‐CoV‐2 infection. Hence, we studied the ex ‐ vivo phenotypic profile of CD4+ and CD8+ memory T cell subsets and the circulating levels of common γc cytokines in COVID‐19 individuals more than 150 days after infection following RT PCR confirmation.
2. MATERIALS AND METHODS
2.1. Ethics statement
The study was approved by the Ethics Committees of ICMR‐NIRT (NIRT‐INo:2020047) and NIE (NIE/IHEC/202008‐01). Informed written consent was obtained from all participants. All the methods were performed in accordance with the relevant institutional ethical committee guidelines.
2.2. Study population
Acute COVID‐19 samples were obtained from individuals following RT‐PCR confirmation (within 15–30 days; n = 46) and convalescent COVID‐19 individual's samples were collected between days 31 and 60, n = 33; 61 and 90, n = 38; 91 and 120, n = 34; 121 and 150, n = 32; 151 and 180, n = 37; and more than 180, n = 40 days post symptom onset, residing in Chennai and Tiruvallur were enrolled in the study between November 2020 and December 2020 after taking informed consent from the enrolled study individuals. 50 , 51 Those who had active COVID‐19 infection under home isolation and recovered COVID‐19 patients within 0–15 days of RT‐PCR confirmation were excluded from the study. The age group ranged between 18 and 75 years. COVID‐19 was confirmed by RT‐PCR in government‐approved laboratories. In brief, nasopharyngeal swabs and oropharyngeal (throat) swabs from individuals suspected of COVID‐19 were obtained by the healthcare provider. RNA isolated and purified from specimens was reverse transcribed to cDNA and amplified. Thermocycling conditions composed of 30 min at 48°C for reverse transcription, 10 min at 95°C for activation of the DNA polymerase, and 45 cycles of 15 s at 95°C and 1 min at 60°C. Fluorescence measurements were taken and the threshold cycle (CT) value for each sample was estimated by determining the point at which fluorescence surpassed a threshold limit set at the mean plus 10 standard deviations beyond the baseline. A test result was calculated positive if two or additional of the SARS genomic targets exhibited positive results (CT < 45 cycles) and all positive and negative control reactions provided an accepted range. Those individuals who did not experience any symptoms during the entire course of illness were considered as asymptomatic and those who required supplemental oxygen support therapy or those who were admitted in ICU for oxygen support were considered as severely ill. Rest was classified under the mild illness category.
2.3. Hematology and flow cytometry
Hematology was performed on all individuals using the Act‐5 Diff hematology analyzer (Beckman Coulter). Demographic details and other clinical parameters are shown in Table 1 decsibed previously. 50 , 51 Whole blood was used for ex‐vivo phenotyping and it was performed on all individuals. Briefly, 250 μL aliquots of whole blood were added to a cocktail of monoclonal antibodies. T cell phenotyping was performed using antibodies directed against CD45‐Peridinin chlorophyll protein (PerCP), CD3− phycoerythrin (PE) Cy7, CD8‐AmCyan, CD28‐allophycocyanin (APC) H7, CD45RA‐Pacific Blue, and CCR7‐FITC and CD95– PE. Naive cells were classified as CD45RA+ CCR7+ CD95– CD28+, central memory cells (TCM) as CD45RA– CCR7+ CD95+ CD28+, effector memory cells (TEM) as CD45RA–CCR7– CD95+ CD28, Terminal effector memory cells (TTEM) as CD45RA– CCR7– CD95+ CD28–, stem cell memory (TSCM) as CD45RA+ CCR7+ CD95+ CD28+, and transitional memory cells (TTM) as CD45RA+ CCR7– CD95+ CD28+. 14 Regulatory T cell phenotyping was performed using CD3 APC‐Cy‐7, CD4 AmyCyan, CD8 PerCP, CD25 APC, CD127 FITC, Foxp3 PE, and regulatory T cells were classified as CD4+ CD25+ Foxp3+ CD127dim. 14 Following 30 min of incubation at room temperature, erythrocytes were lysed using 2 ml of FACS lysing solution (BD Biosciences Pharmingen), cells were washed twice with 2 ml of PBS and suspended in 200 ul of PBS (Lonza, Walkersville, MD). Eight‐color flow cytometry was performed on a FACS Canto II flow cytometer with FACSDIVA software, version 6 (Becton Dickinson). The gating was set by forward and side scatter, and 100 000 gated events were acquired. Gating strategy for memory T cell subsets is shown in Supplementary Figure 1. Data were collected and analyzed using FLOW JO software (TreeStar, Ashland, OR). Leukocytes were gated using CD45 expression versus side scatter. Compensation and gating boundaries were adjusted using unstained, single‐stained, and Fluorescence Minus One (FMO) controls. FMO controls for each marker were used to calculate fluorescence intensity for the population. Total lymphocyte counts were obtained from the hematology profile and the percentage of gated lymphocytes by flow cytometry was used to calculate the absolute numbers of T cell subsets. Absolute counts of lymphocyte populations were calculated based on the equation: Absolute number/mm3 of Leukocytes subset = [percent of lymphocyte × total number of white blood cells per mm3]/100). 15
TABLE 1.
. Demographics and clinical parameters of the study population
| Days after RT‐PCR confirmation | 15–30 days | 31–60 days | 61–90 days | 91–120 days | 121–150 days | 151–180 days | More than 180 days |
|---|---|---|---|---|---|---|---|
| Subjects enrolled | n = 46 | n = 33 | n = 38 | n = 34 | n = 32 | n = 37 | n = 40 |
| Median age (range) | 41.5(18‐70) | 36(25‐68) | 45(19‐59) | 45 (21‐69) | 45.5 (27‐59) | 42 (23‐58) | 38.5(21‐78) |
| Gender (M/F) | 27/19 | 17/18 | 22/15 | 22/12 | 14/18 | 23/16 | 26/14 |
| Mild disease no. (%) | 37 (83) | 30 (73) | 31 (82) | 29 (85) | 27 (84) | 32 (86) | 35 (87) |
| Severe disease no. (%) | 8 (17) | 3 (27) | 7 (18) | 5 (15) | 5 (16) | 5 (14) | 5 (13) |
| Fever, no. (%) | 29 (67) | 22 (65) | 28 (74) | 23 (74) | 25 (83) | 23 (72) | 17 (47) |
| Chills, no. (%) | 9 (21) | 5 (15) | 2 (5) | 7 (22) | 4 (13) | 1 (3) | 3 (8) |
| Cough, no. (%) | 21 (49) | 20 (59) | 14 (37) | 15 (48) | 14 (47) | 17 (53) | 12 (33) |
| Sore throat, no. (%) | 21 (49) | 12 (35) | 11 (29) | 12 (38) | 10 (33) | 16 (50) | 13 (36) |
| Runny nose, no. (%) | 7 (16) | 6 (18) | 5 (13) | NIL | 3 (10) | 6 (19) | 5 (14) |
| Taste loss, no. (%) | 24 (55) | 14 (41) | 17 (44) | 12 (39) | 11 (37) | 20 (63) | 12 (33) |
| Smell loss, no. (%) | 21 (49) | 14 (41) | 21 (55) | 9 (29) | 11 (37) | 16 (50) | 10 (28) |
| Muscle aches, no. (%) | 23 (53) | 20 (59) | 29 (76) | 15 (48) | 18 (60) | 21 (66) | 13 (36) |
| Joint pain, no. (%) | 21 (49) | 18 (53) | 20 (53) | 10 (32) | 18 (60) | 14 (44) | 9 (25) |
| Abdominal pain, no. (%) | 3 (7) | 3 (9) | 4 (11) | 2 (6.5) | 3 (10) | 2 (7) | 3 (8) |
| Vomit, no. (%) | 3 (7) | 4 (12) | 5 (13) | 4 (13) | 3 (10) | 5 (16) | 3 (8) |
| Diarrhea, no. (%) | 10 (23) | 5 (15) | 4 (11) | 4 (13) | 6 (30) | 5 (16) | 2 (6) |
| Seizures, no. (%) | NIL | 1 (3) | NIL | NIL | NIL | NIL | NIL |
| Hypertension, no. (%) | 11 (26) | 7 (21) | 7 (18) | 7 (23) | 9 (30) | 9 (28) | 8 (22) |
| Diabetes, no. (%) | 8 (19) | 7 (21) | 11 (30) | 9 (29) | 11 (37) | 8 (25) | 7 (19) |
| Asthma, no. (%) | 2 (5) | 2 (6) | 1 (3) | 1 (3) | NIL | 1 (3) | NIL |
| Chronic Kidney Disease, no. (%) | NIL | NIL | NIL | NIL | 1 (3) | NIL | 1 (3) |
| Neuro, no. (%) | NIL | NIL | 2 (5) | NIL | NIL | NIL | NIL |
| Heart, no. (%) | 1 (6) | 2 (3) | 1 (3) | NIL | NIL | 1 (3) | NIL |
| Rheumatic fever, no. (%) | NIL | NIL | 1 (3) | NIL | NIL | 1 (3) | NIL |
| Corticosteroids, no. (%) | 4 (9) | 3 (9) | 2 (5) | 3 (10) | 1 (3) | 1 (3) | NIL |
| Antiviral drug, no. (%) | 4 (9) | 5 (15) | 2 (5) | 4 (13) | NIL | NIL | NIL |
| Required hospitalization no. (%) | 10 (22) | 9 (27) | 14 (37) | 17 (50) | 8 (25) | 20 (54) | 27 (68) |
| Required mechanical oxygen support no. (%) | NIL | NIL | NIL | NIL | NIL | NIL | NIL |
2.4. Enzyme‐linked immunosorbent assay
Circulating levels of IL‐2, IL‐7, IL‐15, and IL‐21 were measured using the Duo set ELISA kit (R&D Systems) by ELISA, according to the manufacturer's instructions. The lowest detection limits were as follows: IL‐2, 31.2 pg/ml; IL‐7, 7.813 pg/ml; IL‐15, 16.625 pg/ml; IL‐21, 15.6 pg/ml.
2.5. Statistical analysis
Data analyses were performed using GraphPad PRISM.9 (GraphPad Software, Inc., San Diego, CA, USA). Cross‐sectional analysis of frequency of memory cell subsets and hematology analysis was performed using polynomial model for best‐fit curve (either first‐order or second‐order model). Geometric means (GM) were used for measurements of central tendency. Comparative analysis was done using Kruskal–Wallis test with Dunn's multiple comparisons. Statistically significant differences were analyzed using the nonparametric Mann–Whitney U test used to compare mild versus severe. Stata 15 (College Station, TX) was used to perform the Multiple logistic regression analysis.
3. RESULTS
3.1. Study population characteristics
The study population demographics and clinical characteristics are shown in Table 1 described previously. 50 , 51 (There was no significant difference in age or sex between the study groups).
3.2. Expansion of lymphocyte absolute counts and percentages in convalescent COVID‐19 individuals over a period of time
To examine the absolute numbers and percentages of lymphocytes in acute and convalescent COVID‐19 individuals over time, we examined them in 7 groups of COVID‐19 individuals. Both percentage and absolute counts lymphocytes were shown to increase from days 15–30 till 121–150 days following which the levels plateaued (Supplementary Figure 1.) Analysis was done by first‐order model polynomial model fit curve for lymphocyte absolute count R = 0.25 by Akaike's Information Criterion. Thus, lymphocyte counts increase over time post‐COVID‐19.
3.3. Alterations in frequencies of circulating CD4+ memory T cell subsets in convalescent COVID‐19 individuals over a period of time
To examine the frequencies and distribution of CD4+ T cell memory subsets in convalescent COVID‐19 individuals over time, we evaluated the ex vivo frequencies of memory CD4+ T cell subsets (Naïve cells, central memory, effector memory, transitional memory, terminal effector memory, and stem cell memory T cells) and regulatory T cells in the 7 groups of COVID‐19 individuals. The gating approach is illustrated in Supplementary Figure 2. As shown in Figure 1, cross‐sectional analysis showed that the frequencies of naïve cells started decreasing from days 31–60 and gradually thereafter (first‐order model polynomial model fit curve, R = 0.80 by Akaike's Information Criterion). After 121 days of infection, the frequencies of naïve cells plateaued. Similarly, regulatory T cells started decreasing from days 31–60 (first‐order model polynomial model fit curve, R = 0.88 by Akaike's Information Criterion), then plateaued after 121–150 days. In contrast, the frequencies of central memory (first‐order model polynomial model fit curve, R = 0.18 by Akaike's Information Criterion), effector memory (second‐order model polynomial model fit curve, R = 0.35 by Akaike's Information Criterion), transitional memory (first‐order model polynomial model fit curve, R = 0.59 by Akaike's Information Criterion), terminal effector memory (first‐order model polynomial model fit curve, R = 0.82 by Akaike's Information Criterion) and stem cell memory cells (first‐order model polynomial model fit curve, R = 0.71 by Akaike's Information Criterion) started increasing from days 15–30 till 91–120 days. After 151 days, all the subsets were plateaued. As shown in Supplementary Figure 3, the comparative analysis also exhibited significant differences between various time intervals. CD4+ naïve and regulatory T cells exhibited a significant decrease from day 15–30 till 91–120 days. In contrast, the frequencies of central memory, effector memory, transitional memory, terminal effector memory, and stem cell memory cells started significantly increasing from days 15–30 till 91–120 days. The 95% of confidence intervals were shown in Supplementary Table 1. Thus, CD4+ T cell memory subsets frequencies are altered over time of post COVID‐19 infection.
FIGURE 1.
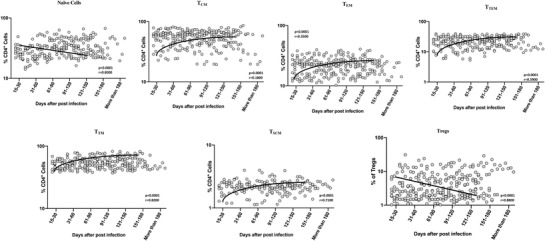
Alterations in frequencies of circulating CD4+ memory T cell subsets in convalescent COVID‐19 individuals over a period of time. Analysis of memory T cell subsets (Naïve cells (TN), central memory cells (TCM), effector memory cells (TEM), Terminal effector memory cells (TTE), stem cell memory (TSCM), transitional memory cells (TTM) and Treg (regulatory T cells) from acute and convalescent COVID‐19 individuals, classified the groups based on days since RT‐PCR confirmation. The frequencies of CD4+ memory T cell subsets are shown with preferred model for best fit curve and each dot represents single individuals. Thick black line represents best fit curve.
3.4. Alterations in frequencies of circulating CD8+ naïve cells and terminal effector memory T cell subsets in convalescent COVID‐19 individuals over a period of time
To examine the frequencies and distribution of CD8+ T cell memory subsets in convalescent COVID‐19 individuals over time, we evaluated the ex vivo frequencies of memory CD8+ T cell subsets (Naïve cells, central memory, effector memory, transitional memory, terminal effector memory, and stem cell memory T cells) in 7 groups of COVID‐19 individuals. The gating strategy is illustrated in Supplementary Figure. 4. As shown in Figure 2, cross‐sectional analysis showed that the frequencies of naïve cells started decreasing from days 31–60 and gradually decreasing (first‐order model polynomial model fit curve, R = 0.84 by Akaike's Information Criterion). After 121 days of infection, the frequencies of naïve cells plateaued. In contrast, the frequencies of terminal effector memory cells (second‐order model polynomial model fit curve, R = 0.44 by Akaike's Information Criterion) started increasing from days 15–30 till 91–120 days and after 151 days plateaued. As shown in Supplementary Figure 5, the comparative analysis also exhibited significant differences between various time intervals. CD8+ naïve T cells exhibited a significant decrease from days 15–30 till 91–120 days. In contrast, the frequencies of transitional memory T cells started significantly increasing from days 15–30 till 91–120 days. The 95% of confidence intervals are shown in Supplementary Table 1. Thus, CD8+ T cell memory subsets frequencies are altered over time following post‐COVID‐19 infection.
FIGURE 2.
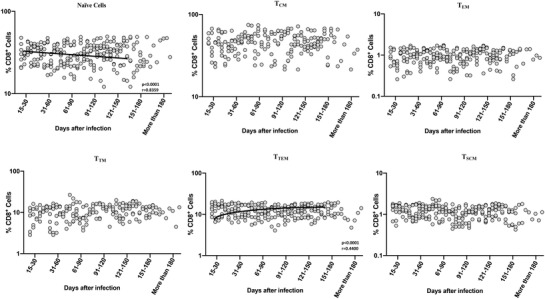
Alterations in frequencies of circulating CD8+ naïve cells and terminal effector memory T cell subsets in convalescent COVID‐19 individuals over a period of time. Analysis of CD8+ memory T cell subsets (naïve cells [TN], central memory cells [TCM], effector memory cells [TEM], Terminal effector memory cells [TTE], stem cell memory [TSCM], transitional memory cells [TTM]) from acute and convalescent COVID‐19 individuals classified the groups based on days since RT‐PCR confirmation. The frequencies of memory T cell subsets are shown with preferred model for best fit curve and each dot represent single individuals. Thick black line represents best fit curve
3.5. Decreased levels of common γ−chain cytokines in convalescent COVID‐19 individuals over a period of time
To estimate the levels of common γ−chain cytokines in convalescent COVID‐19 individuals over time, we determined the plasma levels of IL‐2, IL‐7, IL‐15, and IL‐21 in 7 groups of COVID‐19 individuals. As shown in Figure 3, cross‐sectional analysis exhibited that the levels of IL‐2, IL‐4, IL‐15, and IL‐21 started steadily decreasing from days 15–30, first‐order model polynomial model fit curve, IL‐2, R = 0.064, IL‐7, R = 0.21, IL‐15, R = 0.16, and IL‐21, R = 0.19 by Akaike's Information Criterion) till 150 days after infection. The 95% of confidence intervals are shown in Supplementary Table 1. Thus, plasma levels of common γ−chain cytokines are altered over time following post‐COVID‐19 infection.
FIGURE 3.
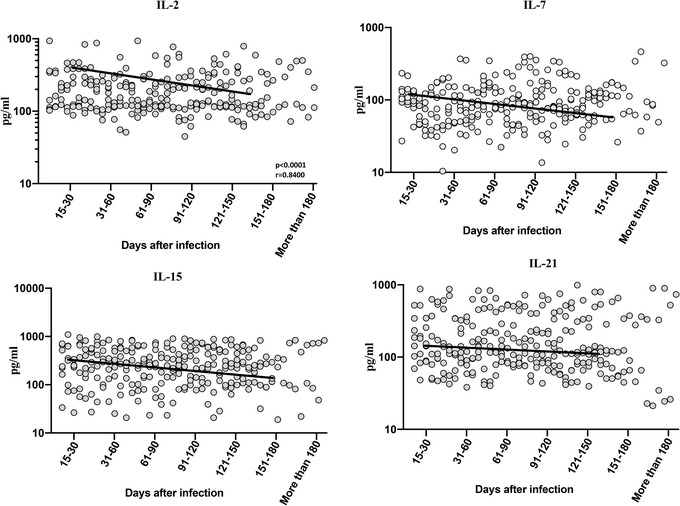
Decreased levels of common γ−chain cytokines in convalescent COVID‐19 individuals over a period of time. (A) Circulating plasma levels of common γ−chain cytokines IL‐2, IL‐7, IL‐15, and IL‐21 from acute and convalescent COVID‐19 individuals classified the groups based on days since RT‐PCR confirmation. The levels of common γ−chain cytokines were shown with preferred model for best fit curve and each dot represent single individuals. Thick black line represents best fit curve
3.6. Severe COVID‐19 disease is associated with altered frequencies CD4+ and CD8+ memory T cell subsets and decreased common γ−chain cytokines
To examine the relationship between lymphocyte counts and disease severity, we determined the absolute counts of lymphocytes in mild and severe COVID‐19 individuals. The study population demographics and clinical characteristics are shown in Table 2. As shown in Figure 4A, lymphocyte absolute count (GM of 2939 cells/μl in mild, 1590 cells/μl in severe, p = 0.0202) and percentage of lymphocytes (GM of 43.06% in mild, 32.36% in severe, p = 0.0090) were significantly lower in severe COVID‐19 compared with mild COVID‐19. Next, we examined the frequencies of CD4+ memory T cell subsets in mild and severely diseased COVID‐19 individuals. As shown in Figure 4B, the frequencies of naïve cells (GM of 27.2% in mild, 51.8% in severe, p < 0.0001) and regulatory T cells (GM of 5.5% in mild, 11.9% in severe, p < 0.0001) were significantly elevated in severe when compared to the mild COVID‐19 patients. In contrast, the frequencies of central memory (GM of 43.5% in mild, 31.0% in severe, p < 0.0001), effector memory (GM of 29.2% in mild, 16.2% in severe, p < 0.0001), and stem cell memory (GM of 2% in mild, 1.6% in severe, p = 0.0126) were significantly lower in severe than the mild COVID‐19 patients. Next, we compared the frequencies of CD8+ memory T cell subsets between mild and severe COVID‐19 patients. As illustrated in Figure 4C, CD8+ memory T cell compartment did not exhibit any significant difference between the 2 groups.
TABLE 2.
Demographics and clinical parameters of the study population
| Days after RT‐PCR confirmation | Mild | Severe |
|---|---|---|
| Subjects enrolled | n = 30 | n = 15 |
| Median age (range) | 39 (18‐81) | 48 (22‐70) |
| Gender (M/F) | 14/16 | 12/3 |
| Fever, no. (%) | 17 (57) | 13 (87) |
| Chills, no. (%) | 4 (13) | 6 (40) |
| Cough, no. (%) | 15 (50) | 7 (47) |
| Sore throat, no. (%) | 16 (53) | 5 (33) |
| Runny nose, no. (%) | 5 (17) | 2 (13) |
| Taste loss, no. (%) | 21 (70) | 5 (33) |
| Smell loss, no. (%) | 17 (57) | 6 (40) |
| Muscle aches, no. (%) | 24 (80) | 9 (60) |
| Joint pain, no. (%) | 18 (60) | 5 (33) |
| Abdominal pain, no. (%) | 2 (7) | 1 (7) |
| Vomit, no. (%) | 2 (7) | 1 (7) |
| Diarrhea, no. (%) | 4 (13) | 6 (40) |
| Seizures, no. (%) | NIL | NIL |
| Hypertension, no. (%) | 7 (23) | 5 (33) |
| Diabetes, no. (%) | 4 (13) | 4 (27) |
| Asthma, no. (%) | 1 (3) | 1 (7) |
| Chronic kidney disease, no. (%) | NIL | NIL |
| Neuro, no. (%) | NIL | NIL |
| Heart, no. (%) | NIL | 1 (7) |
| Rheumatic fever, no. (%) | NIL | NIL |
| Corticosteroids, no. (%) | NIL | 3 ( 20) |
| Antiviral drug, no. (%) | NIL | 4 (27) |
| Required hospitalization no. (%) | NIL | 6 (40) |
| Required mechanical oxygen support no. (%) | NIL | NIL |
FIGURE 4.
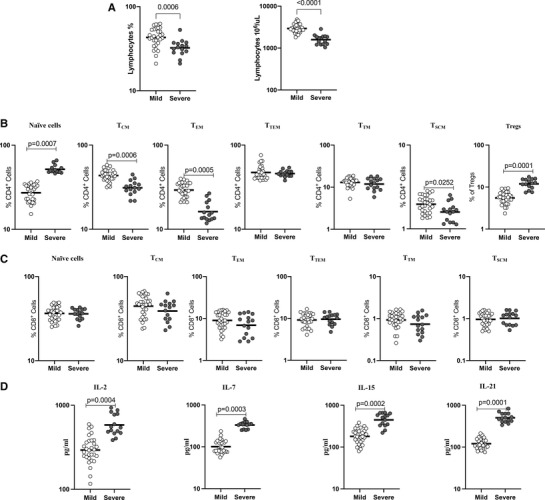
Severe COVID‐19 disease associated with altered frequencies CD4+ and CD8+ memory T cell subsets and decreased common γ−chain cytokines. (A) Lymphocyte absolute count and percentage were shown for mild (n = 30) and severe (n = 15) COVID‐19 individuals sampled between days 15 and 60 following RT‐PCR confirmation. The data are represented as scatter plots with each circle representing a single individual. (B) The frequencies of CD4+ T cell memory subsets in mild (n = 30) and severe (n = 15) COVID‐19 individuals sampled between days 15 and 60 following RT‐PCR confirmation. (C) The frequencies of CD8+ T cell memory subsets in mild (n = 30) and severe (n = 15) COVID‐19 individuals sampled between days 15 and 60 following RT‐PCR confirmation. (D) Circulating plasma levels of common γ−chain cytokines IL‐2, IL‐7, IL‐15, and IL‐21 in mild (n = 30) and severe (n = 15) COVID‐19 sampled between days 15 and 60 following RT‐PCR confirmation. The data are represented as scatter plots with each circle representing a single individual. p‐Values were calculated using the Mann– Whitney U‐test.
In addition, we analyzed the effect of COVID‐19 disease severity on common γc cytokines levels. The levels of IL‐2 (GM of 293.4 pg/ml in mild, 582.3 pg/ml in severe, p < 0.0001), IL‐7 (GM of 100.8 pg/ml in mild, 331.3 pg/ml in severe, p < 0.0001), IL‐15 (GM of 180.3 pg/ml in mild, 446.7 pg/ml in severe, p < 0.0001), and IL‐21 (GM of 120.5 pg/ml in mild, 500.7 pg/ml in severe, p < 0.0001) were significantly higher in severe compared to mild COVID‐19 (Figure 4D). Thus, severe COVID‐19 disease is associated with altered frequencies of memory T cell subsets and diminished levels of common γc cytokines.
3.7. Association between memory T cell subsets and common γc cytokines levels
Next, we wanted to determine the relationship between Memory T cell subsets and common γc cytokines levels in 7 groups of COVID‐19 individuals. As shown in Figure 5A, CD4+ naïve T cells exhibited a significant positive correlation with regulatory T cells. In contrast, other memory subsets showed a significantly negative correlation with regulatory T cells. Further, we performed the correlation analysis between cytokines and memory subsets, CD4+ memory T cell subsets showed significant negative correlation with common γc cytokines levels (Figure 5B). Between CD8+ T cell subsets and common γc cytokines levels, mostly all the subsets with exception of TTEM showed a significant positive correlation with common γc cytokines levels (Figure 5C). Finally, we performed correlation with clinical parameters and memory T cell subsets and common γc cytokines levels. There was no significant correlation among clinical parameters and memory T cell subsets and common γc cytokines levels.
FIGURE 5.
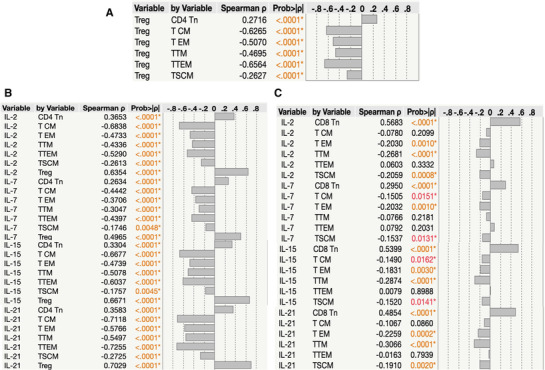
Association between Memory T cell subsets and common γc cytokines levels Multiparametric correlation plot of memory T cell subsets (naïve cells [TN], central memory cells [TCM], effector memory cells [TEM], terminal effector memory cells [TTE], stem cell memory [TSCM], transitional memory cells [TTM]), and common γc cytokines levels (IL‐2, IL‐7, IL‐15, and IL‐21) from all 7 groups of convalescent COVID‐19 individuals classified as groups based on days since RT‐PCR confirmation. Spearman's correlation coefficients are visualized. (A) Correlation analysis between regulatory T cells versus memory CD4+ T cell subsets. (B) Correlation analysis between absolute numbers of memory CD4+ T cell subsets Vs common γc cytokines levels (IL‐2, IL‐7, IL‐15, and IL‐21). (C) Correlation analysis between absolute numbers of memory CD8+ T cell subsets versus common γc cytokines levels (IL‐2, IL‐7, IL‐15, and IL‐21)
3.8. Multivariate logistic model for the disease severity of COVID‐19
Multivariate logistic regression analysis was done to obtain the statistically significant independent determinants of COVID‐19 illness severity. Multivariable logistic regression analysis revealed that diabetes (OR = 3.21, 95% CI: 1.84–9.74; p = 0.002), CD4+ naïve T cells (OR = 0.32, 95% CI: 0.19–0.45; p = 0.031), and regulatory T cells (OR = 0.39, 95% CI: 0.15‐0.67; p = 0.043), the cytokine levels of IL‐7 (OR = 0.967, 95% CI: 0.959‐0.985; p = 0.023) and IL‐21 (OR = 0.969, 95% CI: 0.985 – 1.005; p = 0.013) were independent risk factors associated with severe COVID‐19 (Table 3).
TABLE 3.
Multivariate logistic regression model analysis
| Parameters | OR | 95% CI | p‐Value |
|---|---|---|---|
| Age | 1.021 | 0.963–1.042 | 0.45 |
| Sex Female | 1.000 | 0.346–1.554 | 0.49 |
| Male | 0.775 | ||
| Fever | 0.69 | 0.38–1.24 | 0.36 |
| Chills | 0.997 | 0.937–1.035 | 0.89 |
| Cough | 0.75 | 0.36–1.96 | 0.65 |
| Sore throat | 0.42 | 0.17–1.18 | 0.21 |
| Runny nose | 0.86 | 0.46–1.84 | 0.87 |
| Taste loss | 0.58 | 0.19–2.43 | 0.54 |
| Smell loss | 0.65 | 0.32–1.43 | 0.28 |
| Muscle aches | 1.65 | 0.63–2.74 | 0.31 |
| Joint pain | 1.15 | 0.41–3.73 | 0.84 |
| Abdominal pain | 0.77 | 0.35–1.54 | 0.38 |
| Vomit | 1.58 | 0.69–3.25 | 0.29 |
| Diarrhea | 0.41 | 0.15–1.04 | 0.072 |
| Hypertension | 2.23 | 0.89–3.95 | 0.068 |
| Diabetes | 3.21 | 1.84–9.74 | 0.002 |
| Asthma | 2.32 | 0.72–6.53 | 0.27 |
| CD4+ naïve T cells | 0.32 | 0.19–0.45 | 0.031 |
| Central memory T cells | 0.16 | 0.04–0.38 | 0.26 |
| Effector memory T cells | 0.18 | 0.07–0.32 | 0.30 |
| Transitional memory T cells | 0.66 | 0.56–0.87 | 0.158 |
| Terminal effector memory T cells | 0.45 | 0.25–0.71 | 0.17 |
| Stem cell memory T cells | 0.16 | 0.05–0.36 | 0.18 |
| Regulatory T cells | 0.39 | 0.15–0.67 | 0.043 |
| IL‐2 | 0.997 | 0.956–0.989 | 0.32 |
| IL‐7 | 0.967 | 0.959–0.985 | 0.023 |
| IL‐15 | 0.986 | 0.918 – 1.000 | 0.27 |
| IL‐21 | 0.969 | 0.985 – 1.005 | 0.013 |
4. DISCUSSION
Memory T cells play an important role in viral elimination at the time of re‐infection, however the endurance of SARS‐CoV‐2‐specific memory T cells among COVID‐19 convalescent patients remains vague. In the present study, we performed a precise examination of lymphocytes numbers, memory T cell subset frequencies, and plasma levels of γc cytokines in acute COVID‐19 (defined as days 15–30 from detection) and different groups of convalescent COVID‐19 (7 groups characterized by duration from detection). We have classified convalescent COVID‐19 individuals into different groups based on the duration from RT‐PCR detection (which is the most accurate marker of infection) ranging from 31–60 days to longer than 180 days.
Lymphocyte and their subsets play a vital role in adaptive immune system. 16 A mounting list of reports considering lymphocyte subset counts with patients with COVID‐19 has been explored, many of them concentrating on the predictive importance and relationship of cellular subsets related to disease severity in COVID‐19 patients. 17 , 18 Only a scanty number of studies have focused on the dynamic and longitudinal alterations of lymphocytes, memory T cell subsets and γc cytokines during the course of COVID‐19. Previously, published studies have reported that lymphocytopenia is common during SARS‐CoV‐2 infection, 19 , 20 mainly in severe or critical cases. 21 , 22 Our present study indicates a significant rising trend in the absolute counts and the frequencies of lymphocytes. These findings are consistent with previous studies and they reported that dynamic changes occur after one week of infection and showed an increasing trend in lymphocytes. 23 , 24 , 25 , 26 The existing data implies that peripheral T cell diminution is associated with disease severity and viral load and recovery of counts can happen swiftly either due by subsequent clinical or virological recovery. 27 In consistent with previous published studies, our study also indicates that the percentage and absolute counts were significantly decreased in severe cases in comparison with mild COVID‐19 cases.
Previous data described that SARS‐CoV‐2‐specific memory T cell responses are stimulated following recovery from COVID‐19 disease. Recent data reported SARS‐CoV‐2‐specific memory T cell responses in the initial convalescent period of COVID‐19. 28 , 29 , 30 , 31 , 32 , 33 There is a necessity to understand the kinetics of T cell responses to SARS‐CoV‐2 infection. Since, memory T cells are well‐known to defend against several viral infections. 34 Previous studies reported that 2–4 weeks after COVID‐19 infection, most convalescing patients exhibited enhanced frequency of effector memory‐like CD4+ T cells. 24 , 35 A very recent study reported that CD4+ and CD8+ T cell responses persist for 3–4 months after post symptom onset. 36 Another recent study reported that circulating SARS‐CoV‐2 memory CD4+ and CD8+ T cells are present at ≥6 months post‐symptom onset. 37 Mathew and Chen et al. reported that naïve T cells were decreased in convalescent COVID‐19, but many effector and memory subsets are proportionally increased. 38 , 39 , 40 Very recent data indicated that the proportion of stem cell‐like memory T (TSCM) cells are increased, peaking at ≈120 days post‐symptom onset. 32 Our data indicate that the frequencies of CD4+ central and effector memory cells increased whereas, the frequencies of CD4+ naïve, transitional, stem cell memory T cells, and regulatory cells diminished with increased time of convalescence. Also, the frequencies of CD8+ central memory, effector memory, and terminal effector memory T cells increased, whereas the frequencies of CD8+ naïve cells, transitional memory, and stem cell memory T cells decreased with increased time of convalescence. Thus, our data clearly demonstrate the restoration of T cell subset homeostasis with time in SARS‐CoV2 infection. Treg cells are important for immune homeostasis. It limits the autoimmune effects and constrain inflated inflammatory responses and subsequent viral infections. 41 Circulating CD4+ T‐cells expression of Foxp3 is increased in convalescent COVID‐19 patients in comparison with unexposed individuals. 42 , 43 Similarly, our data also exhibited increased Treg cells in severe patients than the mild/moderate COVID‐19 patients.
The peripheral T cell development, function, and survival require the common γc cytokines 44 and also act as an essential growth factors for T cells. 45 IL‐2 plays a crucial role in T‐cell homeostasis and it is essential for constant expansion of T cell populations. 44 , 45 T‐cell homeostasis, persistence of naïve T‐cell pool requires IL‐7. 46 Severe COVID‐19 patients exhibited elevated serum levels, 19 implies that the IL‐7‐mediated compensatory mechanism is functioning routinely. Besides, it was seen that IL‐2 and IL‐7 levels were enhanced in severe and mild/moderate COVID‐19 patients. 19 , 47 IL‐15 plays a crucial role in supporting the size of the CD8+ T‐cell and memory T‐cell pool 46 and is also involved in T‐cell homeostasis in COVID‐19, although there is a paucity of data for IL‐15 in COVID‐19. Lucas et al. determined that IL‐2, IL‐7, and IL‐15 were enhanced in COVID‐19 and associated with disease severity. 12 , 48 IL‐21 has been shown to support both the cytotoxic and humoral arms of the immune response also act as antiviral response. 49 Hence, our study demonstrates that alterations in the plasma levels of the common γc cytokines, IL‐2, IL‐7, IL‐15, and IL‐21 are correlated with the differential memory T cell compartment modifications seen in acute and convalescent COVID‐19 individuals. Moreover, the multivariate logistic analysis revealed that diabetes, absolute numbers of CD4+ naïve T cells and regulatory T cells, cytokine levels of IL7 and IL‐21 were independent variables associated with severe COVID‐19.
Our study has constraints that we did not examine the functional impact of these alterations in cellular subsets. We have also not explored the persistence of antigen – specific T cell responses in this study. Conversely, it does provide impetus to stimulate the study of the role of these T cell subsets in acute and convalescent COVID‐19 and improve our understanding of memory T cell responses in COVID‐19. In addition, our study underlines the significance of these subsets and the role of common γc cytokines in COVID‐19 infection. Our study has the advantage of a quite large sample size, provides the dynamics of memory T cell subsets and common γc cytokines from early infection to more than 6 months post COVID‐19 infection. Our study also provides the detailed examination of common γc cytokines levels in plasma of IL‐2, IL‐7, IL‐15, and IL‐21 as well as their evolution over time. Our study thus implicates dynamic alterations in memory T cell subsets and common γc cytokines as one of the key events in COVID‐19. The persistence of long‐term SARS‐CoV‐2‐ memory T cells witnessed in our study is indicative of enduring defensive immunity in convalescent COVID‐19 patients.
AUTHORSHIP
S.B., A.R, and N.P.K. designed the study; A.R., N.P.K., A.N., N.S., R.M.R, and V.V. conducted experiments; A.R. and N.P.K. acquired and analyzed data; S.B. and M.M. contributed reagents and also revised subsequent drafts of the manuscript; responsible for the enrolment of the participants and also contributed to acquisition and interpretation of clinical data (M.M., J.W.V.T, CP G.K.); coordinated field operations (M.M., J.W.V.T, M.S.K., T.B., M.P.), coordinated the laboratory processing of samples (CP G.K), coordinated data management (R.S., V.S.,); wrote the manuscript (S.B., A.R.). All authors read and approved the final manuscript.
DISCLOSURE
The authors have declared that no conflict of interest.
Supporting information
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary Table.1. 95% of CI of CD4+ and CD8+ memory T cell subsets and common γc cytokines.
ACKNOWLEDGMENTS
The authors thank the Director of the ICMR‐NIRT for the constant support. The authors thank the data entry operators Mr. K. Jaiganesh and Mr. R. Vigneshwaran, and also all the staff members of the ICER department for the timely help. The authors thank D. Sudha Rani Scientist B, staff nurse: M Beula margrete, Technical Officers C: K. Sathish kumar, Annamma Jose, D Augustine, C. Prabakaran, Technical Officers B: P. Ashok Kumar and laboratory technician: T. Mahesh, S. Kalaivani, M. Sheeba Mary, C. Kanagasivam, Technical Assistant: R. Sivakumar, Project Assistants: Y Radhakrishnaiah, R. Swapna Shinde. This work was supported by the Indian Council of Medical Research (ICMR)‐National Institute for Epidemiology and ICMR‐NIRT‐International Center for Excellence in Research. The funders had no role in study design, data collection, and analysis, decision to publish, or preparation of the manuscript.
Rajamanickam A, Kumar NP, Pandiaraj AN, et al. Characterization of memory T cell subsets and common γ−chain cytokines in convalescent COVID‐19 individuals. Journal of Leukocyte Biology. 2022;112:201–212. 10.1002/JLB.5COVA0721-392RR.
REFERENCES
- 1. Perlman S, Netland J. Coronaviruses post‐SARS: update on replication and pathogenesis. Nat Rev Microbiol. 2009;7:439‐50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Xu Z, Shi L, Wang Y, et al. Pathological findings of COVID‐19 associated with acute respiratory distress syndrome. Lancet Respir Med. 2020;8:420‐422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Guan WJ, Ni ZY, Hu Y, et al, China Medical Treatment Expert Group for, C . Clinical Characteristics of Coronavirus Disease 2019 in China. N Engl J Med. 2020;382:1708‐1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Diao B, Wang C, Tan Y, et al. Reduction and functional exhaustion of t cells in patients with coronavirus disease 2019 (COVID‐19). Front Immunol. 2020;11:827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Song CY, Xu J, He JQ, Lu YQ. Immune dysfunction following COVID‐19, especially in severe patients. Sci Rep. 2020;10:15838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Del Valle DM, Kim‐Schulze S, Huang HH, et al. An inflammatory cytokine signature predicts COVID‐19 severity and survival. Nat Med. 2020;26:1636‐1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Piot P, Larson HJ, O'Brien KL, et al. Immunization: vital progress, unfinished agenda. Nature. 2019;575:119‐129. [DOI] [PubMed] [Google Scholar]
- 8. Raeber ME, Zurbuchen Y, Impellizzieri D, Boyman O. The role of cytokines in T‐cell memory in health and disease. Immunol Rev. 2018;283:176‐193. [DOI] [PubMed] [Google Scholar]
- 9. Gu XX, Yue FY, Kovacs CM, Ostrowski MA. The role of cytokines which signal through the common gamma chain cytokine receptor in the reversal of HIV specific CD4(+) and CD8(+) T cell anergy. PLoS One. 2007;2:e300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ferrari G, King K, Rathbun K, et al. IL‐7 enhancement of antigen‐driven activation/expansion of HIV‐1‐specific cytotoxic T lymphocyte precursors (CTLp). Clin Exp Immunol. 1995;101:239‐48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schluns KS, Lefrancois L. Cytokine control of memory T‐cell development and survival. Nat Rev Immunol. 2003;3:269‐79. [DOI] [PubMed] [Google Scholar]
- 12. Lucas C, Wong P, Klein J, et al. Longitudinal analyses reveal immunological misfiring in severe COVID‐19. Nature. 2020;584:463‐469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Arenas‐Ramirez N, Zou C, Popp S, et al. Improved cancer immunotherapy by a CD25‐mimobody conferring selectivity to human interleukin‐2. Sci Transl Med. 2016;8:367ra166. [DOI] [PubMed] [Google Scholar]
- 14. Maecker HT, McCoy JP, Nussenblatt R. Standardizing immunophenotyping for the Human Immunology Project. Nat Rev Immunol. 2012;12:191‐200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rajamanickam A, Munisankar S, Bhootra Y, et al. Altered levels of memory T cell subsets and common gammac cytokines in Strongyloides stercoralis infection and partial reversal following anthelmintic treatment. PLoS Negl Trop Dis. 2018;12:e0006481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cossarizza A, De Biasi S, Guaraldi G, Girardis M, Mussini C. Modena Covid‐19 Working, G. (2020) SARS‐CoV‐2, the Virus that Causes COVID‐19: cytometry and the new challenge for global health. Cytometry A;97:340‐343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liu Z, Long W, Tu M, et al. Lymphocyte subset (CD4+, CD8+) counts reflect the severity of infection and predict the clinical outcomes in patients with COVID‐19. J Infect. 2020;81:318‐356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Huang W, Berube J, McNamara M, et al. Lymphocyte subset counts in COVID‐19 patients: a meta‐analysis. Cytometry A. 2020;97:772‐776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497‐506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang D, Hu B, Hu C, et al. Clinical characteristics of 138 hospitalized patients with 2019 Novel Coronavirus‐Infected Pneumonia in Wuhan, China. 2020:1061‐1069. [DOI] [PMC free article] [PubMed]
- 21. Tan L, Wang Q, Zhang D, et al. Lymphopenia predicts disease severity of COVID‐19: a descriptive and predictive study. Signal Transduct Target Ther. 2020;5:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhao Q, Meng M, Kumar R, et al. Lymphopenia is associated with severe coronavirus disease 2019 (COVID‐19) infections: a systemic review and meta‐analysis. Int J Infect Dis. 2020;96:131‐135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Moratto D, Chiarini M, Giustini V, et al. Flow cytometry identifies risk factors and dynamic changes in patients with COVID‐19. J Clin Immunol. 2020;40:970‐973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Weiskopf D, Schmitz KS, Raadsen MP, et al. Phenotype and kinetics of SARS‐CoV‐2‐specific T cells in COVID‐19 patients with acute respiratory distress syndrome. Sci Immunol. 2020;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Deng Z, Zhang M, Zhu T, et al. Dynamic changes in peripheral blood lymphocyte subsets in adult patients with COVID‐19. Int J Infect Dis. 2020;98:353‐358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rezaei M, Marjani M, Mahmoudi S, Mortaz E, Mansouri D. Dynamic Changes of Lymphocyte Subsets in the Course of COVID‐19. Int Arch Allergy Immunol. 2021;182:254‐262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shrotri M, van Schalkwyk MCI, Post N, et al. 2021) T cell response to SARS‐CoV‐2 infection in humans: a systematic review. PLoS One;16:e0245532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Grifoni A, Weiskopf D, Ramirez SI, et al. Targets of T Cell Responses to SARS‐CoV‐2 Coronavirus in Humans with COVID‐19 Disease and Unexposed Individuals. Cell. 2020;181:1489‐1501 e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Peng Y, Mentzer AJ, Liu G, et al, Oxford Immunology Network Covid‐19 Response, T. c. C., Investigators, I. C., Cornall, R. J., Conlon, C. P., Klenerman, P., Screaton, G. R., Mongkolsapaya, J., McMichael, A., Knight, J. C., Ogg, G., Dong, T . Broad and strong memory CD4(+) and CD8(+) T cells induced by SARS‐CoV‐2 in UK convalescent individuals following COVID‐19. Nat Immunol. 2020;21:1336‐1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sekine T, Perez‐Potti A, Rivera‐Ballesteros O, et al. Robust T Cell Immunity in convalescent individuals with asymptomatic or mild COVID‐19. Cell. 2020;183:158‐168 e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rodda LB, Netland J, Shehata L, et al. Functional SARS‐CoV‐2‐Specific immune memory persists after mild COVID‐19. Cell. 2021;184:169‐183 e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jung JH, Rha MS, Sa M, et al. SARS‐CoV‐2‐specific T cell memory is sustained in COVID‐19 convalescent patients for 10 months with successful development of stem cell‐like memory T cells. Nat Commun. 2021;12:4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sherina N, Piralla A, Du L, et al. Persistence of SARS‐CoV‐2‐specific B and T cell responses in convalescent COVID‐19 patients 6–8 months after the infection. Med (N Y). 2021;2:281‐295 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ahmed R, Gray D. Immunological memory and protective immunity: understanding their relation. Science. 1996;272:54‐60. [DOI] [PubMed] [Google Scholar]
- 35. Gong F, Dai Y, Zheng T, et al. Peripheral CD4+ T cell subsets and antibody response in COVID‐19 convalescent individuals. J Clin Invest. 2020;130:6588‐6599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jiang XL, Wang GL, Zhao XN, et al. Lasting antibody and T cell responses to SARS‐CoV‐2 in COVID‐19 patients three months after infection. Nat Commun. 2021;12:897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dan JM, Mateus J, Kato Y, et al. Immunological memory to SARS‐CoV‐2 assessed for up to 8 months after infection. Science. 2021;371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hanna SJ, Codd AS, Gea‐Mallorqui E, et al. T cell phenotypes in COVID‐19 ‐ a living review. Oxf Open Immunol. 2021;2:iqaa007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mathew D, Giles JR, Baxter AE, et al. Deep immune profiling of COVID‐19 patients reveals distinct immunotypes with therapeutic implications. Science. 2020;369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chen Z, John Wherry E. T cell responses in patients with COVID‐19. Nat Rev Immunol. 2020;20:529‐536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sakaguchi S, Yamaguchi T, Nomura T, Ono M. Regulatory T cells and immune tolerance. Cell. 2008;133:775‐87. [DOI] [PubMed] [Google Scholar]
- 42. Kalfaoglu B, Almeida‐Santos J, Tye CA, Satou Y, Ono M. 2021) T‐cell dysregulation in COVID‐19. Biochem Biophys Res Commun;538:204‐210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jingyi Yang EZ, Maohua Zhong, Qingyu Yang, et al, (2020) Impaired T cell functions along with elevated activated Tregs at the early stage of asymptomatic SARS‐CoV‐2 infection. medRxiv.
- 44. D'Souza WN, Lefrancois L. Frontline: an in‐depth evaluation of the production of IL‐2 by antigen‐specific CD8 T cells in vivo. Eur J Immunol. 2004;34:2977‐85. [DOI] [PubMed] [Google Scholar]
- 45. Geginat J, Sallusto F, Lanzavecchia A. Cytokine‐driven proliferation and differentiation of human naive, central memory, and effector memory CD4(+) T cells. J Exp Med. 2001;194:1711‐9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Surh CD, Sprent J. Homeostasis of naive and memory T cells. Immunity. 2008;29:848‐62. [DOI] [PubMed] [Google Scholar]
- 47. Zheng HY, Zhang M, Yang CX, et al. Elevated exhaustion levels and reduced functional diversity of T cells in peripheral blood may predict severe progression in COVID‐19 patients. Cell Mol Immunol. 2020;17:541‐543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Fara A, Mitrev Z, Rosalia RA, Assas BM. Cytokine storm and COVID‐19: a chronicle of pro‐inflammatory cytokines. Open Biol. 2020;10:200160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wilz SW. A clinical trial of IL‐15 and IL‐21 combination therapy for COVID‐19 is warranted. Cytokine Growth Factor Rev. 2021;58:49‐50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Recovery of Memory B‐cell Subsets and Persistence of Antibodies in Convalescent COVID‐19 Patients. Rajamanickam A, Kumar NP, Nancy P A, Selvaraj N, Munisankar S, Renji RM, V V, Murhekar M, Thangaraj JWV, Kumar MS, Kumar CPG, Bhatnagar T, Ponnaiah M, Sabarinathan R, Kumar VS, Babu S. Recovery of Memory B‐cell Subsets and Persistence of Antibodies in Convalescent COVID‐19 Patients. Am J Trop Med Hyg. 2021 Sep 27;105(5):1255‐1260. doi: 10.4269/ajtmh.21‐0883. PMID: 34583334; PMCID: PMC8592221. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Monocyte numbers, subset frequencies and activation markers in acute and convalescent COVID‐19 individuals. Rajamanickam A, Kumar NP, Pandiarajan AN, Selvaraj N, Munisankar S, Renji RM, Venkatramani V, Murhekar M, Thangaraj JWV, Kumar MS, Kumar CPG, Bhatnagar T, Ponnaiah M, Sabarinathan R, Saravanakumar V, Babu S. Dynamic alterations in monocyte numbers, subset frequencies and activation markers in acute and convalescent COVID‐19 individuals. Sci Rep. 2021 Oct 12;11(1):20254. doi: 10.1038/s41598‐021‐99705‐y. PMID: 34642411; PMCID: PMC8511073. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary Table.1. 95% of CI of CD4+ and CD8+ memory T cell subsets and common γc cytokines.


