Figure 3.
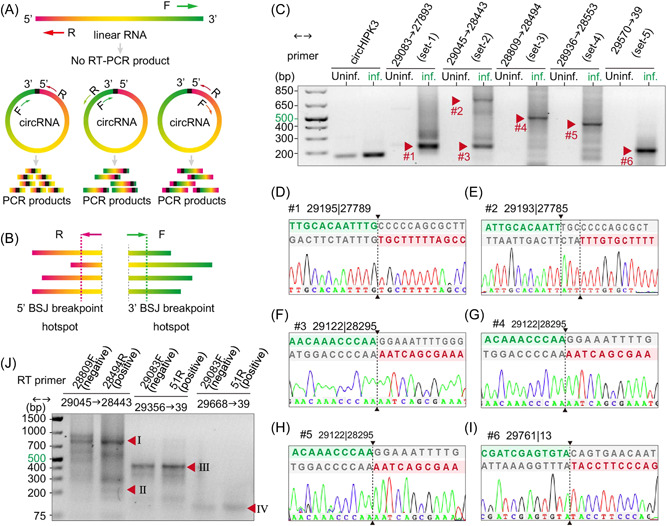
Experimental identification of severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) circular RNAs (circRNAs) in Vero E6 cells. (A) Illustration of inverse reverse‐transcription polymerase chain reaction (RT‐PCR) with divergent primers would selectively amplify different regions of circRNAs but not linear RNAs. (B) Schematic diagram showing divergent primers were designed to amplify as much predicted back‐splice junctions (BSJs) in a given hotspot as possible. (C–I) Inverse RT‐PCR result with selected primer sets were shown in C. Bands indicated by red arrows were sequenced. Representative Sanger sequencing results were shown in D–I. BSJ breakpoints were indicated by dashed lines. Donor (green) and acceptor (red) sequences and downstream/upstream sequences (gray) flanking the junction were aligned to the BSJ sequence. (J) Strand‐specific RT inverse PCR result. Bands corresponding to BSJ 29122 | 28295 (225 bp), BSJ 29195 | 27789 (804 bp), and BSJ 29761 | 13 (431 and 119 bp) were indicated by red arrowheads.
