Figure 5.
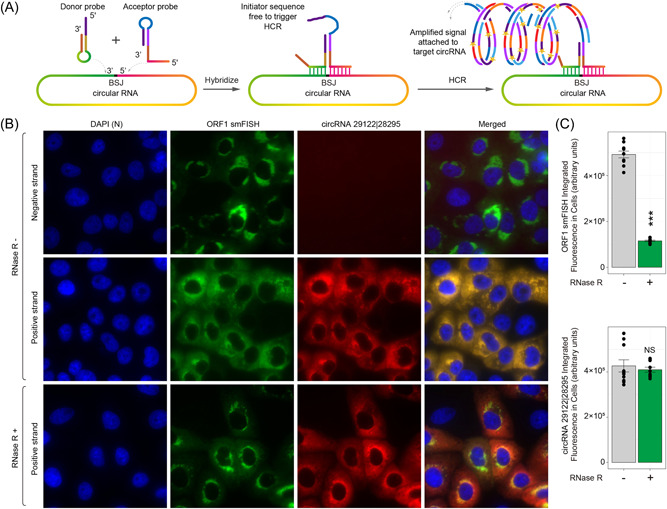
Localization of severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) circular RNAs (circRNAs) in infected Vero E6 cells. (A) Design of amplified fluorescence in situ hybridaization (AmpFISH) probes for the detection of a juxtaposed pair of sequences in the circRNA 29122 | 28295. Only when the two sequences are next to each other, which occurs in the circular RNA but not in linear RNA, the donor and acceptor probes can interact with each other and give rise to an amplified hybridization chain reaction (HCR) signal. (B) Multiplex imaging of the linear RNA corresponding to ORF1 obtained by single‐moleclule FISH (smFISH; green) and circRNA 29122 | 28295 (green) obtained from AmpFISH in the same set of cells. Nuclei were stained with 4′,6‐diamidino‐2‐phenylindole (DAPI). RNAse R treatment is expected to degrade linear but not the circular forms of RNAs. (C) Mean fluorescence intensity in single infected cells that were either treated with RNAse R (green) or left untreated (gray). Linear RNA was detected using ORF1‐specific smFISH probe sets (top) and the circRNA was detected by back‐splice junction (BSJ)‐specific AmpFISH probes pair (bottom).
