Abstract
Through active surveillance and contact tracing from outpatients, we aimed to identify and characterize SARS‐CoV‐2 variants circulating in Porto Velho‐Rondônia, a city in the Brazilian Amazon. As part of a prospective cohort, we gathered information from 2,506 individuals among COVID‐19 patients and household contacts. Epidemiological data, nasopharyngeal swabs, and blood samples were collected from all participants. Nasopharyngeal swabs were tested for antigen rapid diagnostic test and reverse transcription‐polymerase chain reaction (RT‐PCR) followed by genomic sequencing. Blood samples underwent ELISA testing for IgA, IgG, and IgM antibody levels. From 757 specimens sequenced, three were identified as Mu variant, none of the individuals carrying this variant had a travel history in the previous 15 days before diagnosis. One case was asymptomatic and two presented mild symptoms. Two infected individuals from different households caring viruses with additional amino acid substitutions ORF7a P45L and ORF1a T1055A compared to the Mu virus reference sequence. One patient presented IgG levels. Our results highlight that genomic surveillance for SARS‐CoV‐2 variants can assist in detecting the emergency of SARS‐CoV‐2 variants in the community, before its identification in other parts of the country.
Keywords: Active surveillance, Brazilian Amazon, Mu variant, SARS‐CoV‐2
1. INTRODUCTION
Monitoring detection of SARS‐CoV‐2 variants of interest (VOI) across geographic regions provides information on VOIs spread and may aid early identification and characterization of variants of concern (VOC). The SARS‐CoV‐2 Mu variant (Pango lineage B.1.621), first reported in Colombia in January 2021, 1 was classified as a VOI in August 2021 by the World Health Organization (WHO). The Mu variant is characterized by mutations in the gene encoding the SARS‐CoV‐2 spike (S) protein T95I, Y144S, Y145N, 146N insertion, R346K, E484K, N501Y, D614G, P681H, and D950N previously associated with decreased antibody responses and increased transmissibility. 2 , 3 , 4 Many of those mutations (T95I, E484K, N501Y, D614G, P681H, and D950) are also present in Gamma and Delta VOCs. 5 Although the majority of the cases have been reported in South America and the Caribbean, the Mu variant has also been identified in North America, Europe, and Asia. 6
Brazil has the highest number of reported cases of coronavirus disease 2019 (COVID‐19) in South America since the beginning of the pandemic (followed by Argentina and Colombia) and the third‐highest number of reported cases worldwide (behind the United States and India; last access February 2022—https://www.worldometers.info/coronavirus). The North region of Brazil has been the epicenter of multiple epidemic waves, especially the states of Amazonas, Pará, and Rondônia. 7 The SARS‐CoV‐2 Gamma (P.1) variant was first identified in Manaus, the capital of Amazonas, in mild November 2020. 8 From April to June 2021, as part of a household transmission study of the Gamma (P.1) variant, active surveillance for SARS‐CoV‐2 variants was conducted in Porto Velho‐Rondônia, a city in the Brazilian Amazon. Individuals seeking care for COVID‐19 in outpatient facilities and household contacts were screened for SARS‐CoV‐2 infection. Here, we describe cases infected with SARS‐CoV‐2 Mu variant identified among persons who had no history of travel and who lived in Porto Velho, the capital of Rondônia state in western Brazil.
2. PATIENTS AND METHODS
This activity was reviewed by CDC and was conducted consistent with applicable federal law and CDC policy and was approved by the Research Ethics Committee of the Centro de Pesquisa em Medicina Tropical de Rondônia‐CEPEM/RO and by the Brazilian Ethical Committee (Comissão Nacional de Ética em Pesquisa, CONEP: 4.959.645). From April to June 2021, we enrolled patients from 10 outpatient health units designated by local health authorities as COVID‐19 of Porto Velho‐Rondônia, Brazil referral centers as part of a prospective cohort study aimed at characterizing the infection profile of viral variants circulating in important epicenters of SARS‐CoV‐2 transmission. Initially, individuals that provided informed consent were interviewed by study staff and nasal swab specimens were tested with the use of rapid diagnostic tests (RDT) from BinaxNOW COVID‐19 Ag Card (Abbott, USA). From enrolled patients who tested positive by RDT, we collected nasopharyngeal specimens for blood specimens. We also visited the household to collect further epidemiological data, nasal and nasopharyngeal swabs, and blood specimens from all individuals. We followed index case‐patients and household contacts for 14 days. All nasopharyngeal specimens underwent reverse transcription‐polymerase chain reaction (RT‐PCR) using Allplex 2019‐nCoV assay (Seegene) 9 designed for amplifying three viral targets: the E (specific of the subgenus Sarbecovirus), the N and the RdRP genes (both specific to SARS‐CoV‐2) done at Rondônia State Central Public Health Laboratory (LACEN). Viral load was determined using 5 μL of this extracted viral RNA from 140 µL of pooled Swab samples using the QIAamp® Viral RNA Mini Kit (QIAGEN), and the Multiplex One‐Step RT‐qPCR assay for quantification of SARS‐CoV‐2 10 performed at Fundação Oswaldo Cruz (Fiocruz), Rondônia. Samples with cycle threshold (Ct) values <30 for N gene target were transferred in dry ice to Fiocruz, Amazonas, where nucleotide sequencing was performed using Illumina MiSeq or NextSeq platform and the COVIDSEQ Kit (Illumina). 11 Genomic sequences were analyzed using Nextclade software v.1.5.4 (http://clades.nettrain.org) and Pangolin COVID‐19 Linea Assigner v.2.1.7 (https://pangolin.cog-uk.io). Blood specimens were tested for anti‐SARS‐CoV‐2 spike and nucleocapsid‐specific IgM, IgA using ELISA (IgM+A) from Vircell(Spain) 12 and anti‐SARS‐CoV‐2 spike IgG antibodies using Anti‐SARS‐CoV‐2 QuantiVac ELISA (IgG) from Euroimmun 13 according to the manufacturer's instructions at Fiocruz, Rondônia.
3. RESULTS
Among 2506 individuals enrolled and initially tested by RDT, 927 were positive for SARS‐CoV‐2 infection by RT‐qPCR. Complete genome sequencing was performed on 757 samples with Ct < 30. As expected, 694 (92%) specimens tested presented the Gamma variant, the dominant VOC in the Amazon region. Interestingly, we detected, for the first time, three individuals infected with Mu variant in Porto Velho with no travel history prior to the symptoms. Below we describe the epidemiologic and clinical characteristics of the cases identified.
3.1. Case 1
The first SARS‐CoV‐2 Mu variant was detected in an asymptomatic 23‐year‐old male from a respiratory swab collected on June 4, 2021. On the same day, the patient's father tested positive by SARS‐CoV‐2 RDT and RT‐PCR through active surveillance. SARS‐CoV‐2 sequencing from the father's nasopharyngeal swab identified the Gamma variant (Pango lineage P.1). Two household contacts were identified; respiratory specimen from one contact tested negative for SARS‐CoV‐2 while the second one tested positive by SARS‐CoV‐2 RT‐qPCR with a cycle threshold value (Ct) of 30, estimated viral load of 104.3 copies/mL. 10 Genetic sequencing identified SARS‐CoV‐2 Mu variant virus, with additional amino acid substitutions ORF7a P45L and ORF1a T1055A compared to the Mu virus reference sequence (Figure 1 and Table 1). The index case had no underlying medical conditions, no prior history of COVID‐19 vaccination, and no travel in the previous 14 days. Serologic testing of blood specimens collected June 4 tested negative for anti‐SARS‐CoV‐2 IgM, IgA, and IgG antibodies. The patient presented with fever on June 12 and remained RT‐qPCR positive on follow‐up testing on June 18, Ct of 31, and estimated viral load of 103.97 copies/mL.
Figure 1.
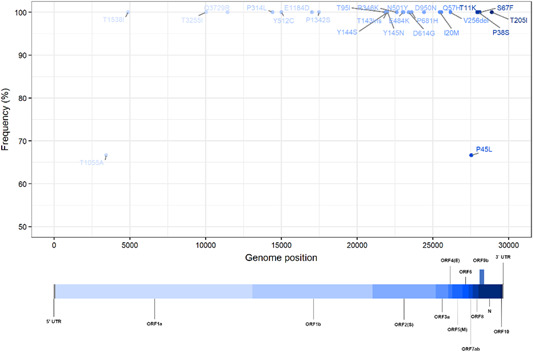
Representation of the genome‐wide mutation frequency of the Mu variant identified in this study. All three cases described here had the defining mutations of the Mu strain, additionally, Cases 1 and 3 had two amino acid substitutions ORF7a P45L and ORF1a T1055A compared to the Mu variant reference sequence
Table 1.
Representation of the genome‐wide mutation frequency of the Mu variant identified in all three cases described with the defining mutations of the Mu strain, compared to the Mu variant reference sequence
| Sequence | Accession ID | Similarity(%) | Nucleotide substitutions | Nucleotide deletions | Nucleotide insertions | Missing (N's) | Length |
|---|---|---|---|---|---|---|---|
| Case 1 | EPI_ISL_10115640 | 99 | 35 | 4 | 3 | 0 | 29 812 |
| Case 2 | EPI_ISL_10115642 | 96 | 32 | 4 | 3 | 957 | 29 646 |
| Case 3 | EPI_ISL_10115641 | 98 | 34 | 4 | 3 | 343 | 29 805 |
Similarity with hCoV‐19/Wuhan/Hu‐1/2019 strain (GISAID accession number EPI_ISL_402125).
3.2. Case 2
The second SARS‐CoV‐2 Mu variant infection was identified in a symptomatic 34‐year‐old male with no travel in the past 14 days and no known contact with Patient 1. The patient presented at an urgent care center on June 18, 2021, with a history of cough, fatigue, myalgia, nausea/vomiting, and retro‐orbital pain for 5 days and tested positive by SARS‐CoV‐2 by RDT. The patient had no underlying medical conditions and had not received the COVID‐19 vaccine. A nasopharyngeal swab tested positive by RT‐qPCR with a Ct of 20, estimated viral load of 107.4 copies/ml. Genetic sequencing identified Mu variant virus infection; however, amino acid substitutions observed in Case 1 were not detected (Figure 1 and Table 1). Serologic testing of blood specimens collected June 18 tested negative by SARS‐CoV‐2 antigen‐specific IgM, IgA, and IgG assays. This patient remained RT‐PCR positive on follow‐up testing on July 1 with a Ct of 24, estimated viral load of 106.18 copies/mL.
3.3. Case 3
A nasopharyngeal specimen collected June 18, 2021, from the mother of Case 2 tested positive by SARS‐CoV‐2 RT‐qPCR with Ct of 27 and an estimated viral load of 105.2 copies/mL. The patient was a 58‐year‐old, healthy female with no underlying medical conditions who complained of cough with onset 2 days prior but had no other symptoms and tested negative by SARS‐CoV‐2 RDT. She had received one dose of the AZD1222 (Oxford‐AstraZeneca) COVID‐19 vaccine on May 11, 2021. Genetic sequencing identified Mu variant virus; the same two amino acid substitutions detected in sequence from Case 1 were present (Figure 1 and Table 1). Serologic testing of blood specimens collected June 18 tested positive for anti‐SARS‐CoV‐2 spike protein 1 (S1) IgG antibody (54.6 relative units/mL) and negative IgM and IgA antibodies. This patient was RT‐PCR positive with a Ct of 27 on July 1 with viral load by RT‐qPCR of 105.19 copies/mL.
Active surveillance and sample collection finished on June 30th, after that no more cases of Mu variant were identified. However, in addition to Mu and Gamma variant viruses, cases of the Delta variant (Pango lineage B.1.617.2) were identified in Porto Velho in June 2021 (Figure 2).
Figure 2.
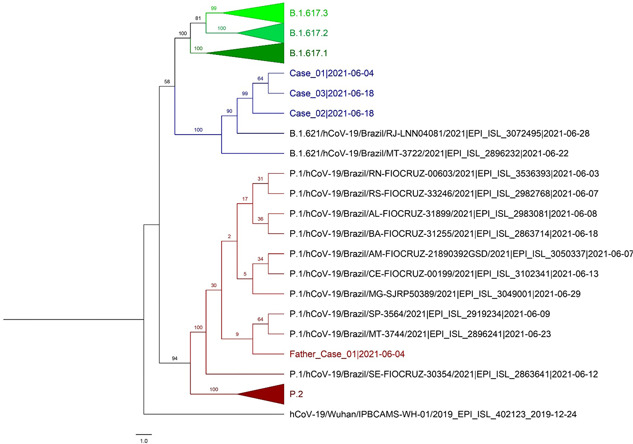
Phylogenetic tree including SARS‐CoV‐2 variant virus sequences identified in Rondônia state, western Brazil, during June–August 2021 (in color): Delta (B.1.617.2, n = 3 [green]), Mu (B.1.621, n = 3 [described here, blue]), and Gamma (P.1, father [red]) lineages. For comparison, 16 sequences obtained from GISAID (www.gisaid.org) for SARS‐CoV‐2 viruses from Delta, Mu, and Gamma lineages identified in the state are shown by genetic distance using a bootstrap analysis to estimate the confidence of the branches
4. DISCUSSION
Following a major international soccer event in Brazil in July 2021, reports of Mu variant viruses in multiple states suggested that international travel might have contributed to the introduction and spread of this variant. We describe three SARS‐CoV‐2 Mu variant infections among persons with no history of travel in Porto Velho, the capital of Rondônia state in western the Brazilian Amazon in June 2021, days before the international event. Interestingly, after the identification of these three cases of Mu variant in Porto Velho new cases of this variant were detected in other parts of Brazil, 14 during the international soccer event. Due to the detection of these variants in very few cases within mild symptoms to asymptomatic in the community, we believe this variant was already circulating in the country with very low frequency before the sports event. As of November 2021, only 21 sequences of My variant from Brazil have been deposited on the GISAID (https://www.gisaid.org).
Our effort and findings highlight the importance of genomic surveillance as part of active epidemiologic surveillance as it allowed for the identification of the Mu variant in the community among individuals who might normally go undetected. Interestingly, the first detection of Mu variant infection occurred in close contact with an individual infected with the Gamma variant, suggesting co‐circulation of Mu VOI and Gamma VOC in the community as early as the first week of June 2021. Mu variant infected cases have not increased in the community after the detection of the three cases described here. We hypothesize that the emergence or a potentially more transmissible VOC (Delta) in the same period in combination with a stationary state of low endemic viral community transmission could be responsible for the limited spread of Mu variant in the Porto Velho community. Lineage replacement of VOCs is a recurrent phenomenon in the local evolution of SARS‐CoV‐2 virus in the Amazon region driven by ecological and virological factors 11 and has been followed among genetic surveillance in different parts of the world.
Our work has some limitations. First, we identified patients at outpatient facilities, which might not seamlessly reflect community transmission patterns. Second, genomic sequence success depends on the high viral load that could be impacted by the timing of sample collection. Third, only Case 3 had measurable SARS‐CoV‐2 specific antibody by serologic assay, and only for IgG. The timing of infection and sample collection as well assay sensitivity could have impacted antibody detection. The kinetics of specific immunoglobulin production against spike‐1 receptor‐binding domain protein shows that most patients produce detectable IgG and IgA/IgM antibodies within 2–3 weeks from the onset of symptoms, while the last blood specimens in this study were collected 14 days after a positive RDT test.
Global surveillance for SARS‐CoV‐2 variants is essential for detecting and tracking disease spread, informing mitigation strategies. However, in resource‐limited contexts, genomic characterization may not be widely feasible but can be implemented in selected reference laboratories. Furthermore, variant genomic surveillance often focuses on hospitalized patients, which often detects new variant viruses associated with more severe illnesses. Expanding active surveillance to include variants from community settings can allow for early detection of emerging SARS‐CoV‐2 variants and provide better insight into the timing of their introduction into new regions.
BRAZIL INVESTIGATION TEAM
Jackson A. S. Queiroz (Laboratório de Virologia Molecular, Fundação Oswaldo Cruz, Fiocruz, Rondônia, Brazil) and Fernanda C. Lessa (Centers for Diseases Control and Prevention (CDC), COVID‐19 Response, Atlanta, Georgia, USA).
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
AUTHOR CONTRIBUTIONS
Conceptualization and validation: Deusilene S. Vieira Dall'Acqua, Luciana Silva‐Flannery, Gabriella S. Oliveira, Charlene Siza, and Mateusz Plucinski. Methodology: Deusilene S. Vieira Dall'Acqua, Felipe G. Naveca, Gabriella S. Oliveira, Ana M. P. Silva, Tárcio P. Roca, Soraya S. Pereira, Juan M. V. Salcedo, Jackson A. S. Queiroz, and Dhelio B. Pereira. Investigation: Juliana F. Silva, Charlene Siza, Mateusz Plucinski, Gabriella S. Oliveira, Luciana Silva‐Flannery, Deusilene S. Vieira Dall'Acqua, Dhelio B. Pereira, Juan M. V. Salcedo, Juliette Morgan, Roberto J. Esteves, Barbara J. Marston, Jackson A. S. Queiroz, and Fernanda C. Lessa. Writing–original draft: Gabriella S. Oliveira, Luciana Silva‐Flannery, Deusilene S. Vieira Dall'Acqua, Charlene Siza, and Tárcio P. Roca. Writing – review and editing: Gabriella S. Oliveira, Luciana Silva‐Flannery, Tárcio P. Roca, and Deusilene S. Vieira Dall'Acqua. Supervision: Juliette Morgan, Roberto J. Esteves, Barbara J. Marston, and Mateusz Plucinski. Software: Tárcio P. Roca, Gabriella S. Oliveira, and Felipe G. Naveca. Funding acquisition: Juliette Morgan, Roberto J. Esteves, and Barbara J. Marston. All authors approved the final version.
ACKNOWLEDGMENTS
The authors thank the Laboratório Central de Saúde Pública de Rondônia (LACEN) for the initial sample processing, and the support provided by the Fiocruz COVID‐19 Genomic Surveillance Network (http://www.genomahcov.fiocruz.br) and the Respiratory Viruses Genomic Surveillance Network of the Brazilian Ministry of Health Central Laboratory Coordination (Coordenação Geral de Laboratórios de Saúde Pública CGLAB). The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention (CDC). This study was supported by CDC/USA.
Oliveira GS, Silva‐Flannery L, Silva JF, et al. Active surveillance and early detection of community transmission of SARS‐CoV‐2 Mu variant (B.1.621) in the Brazilian Amazon. J Med Virol. 2022;94:3410‐3415. 10.1002/jmv.27686
Gabriella Sgorlon Oliveira and Luciana Silva‐Flannery share the first authorship for this paper.
DATA AVAILABILITY STATEMENT
All SARS‐CoV‐2 genomes generated and analyzed in this study are available at the EpiCoV database in GISAID (https://www.gisaid.org) with accession IDs EPI_ISL_10115640, EPI_ISL_10115641, and EPI_ISL_10115642.
REFERENCES
- 1. Laiton‐Donato K, Franco‐Munoz C, Alvarez‐Diaz DA, et al. Characterization of the emerging B.1.621 variant of interest of SARS‐CoV‐2. Infect Genet Evol. 2021;95:105038. 10.1016/J.MEEGID.2021.105038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rees‐Spear C, Muir L, Griffith SA, et al. The effect of spike mutations on SARS‐CoV‐2 neutralization. Cell Rep. 2021;34(12):108890. 10.1016/J.CELREP.2021.108890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Davies NG, Abbott S, Barnard RC, et al. Estimated transmissibility and impact of SARS‐CoV‐2 lineage B.1.1.7 in England. Science. 2021;372(6538):283. 10.1126/science.abg3055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jeyanathan M, Afkhami S, Smaill F, Miller MS, Lichty BD, Xing Z. Immunological considerations for COVID‐19 vaccine strategies. Nat Rev Immunol. 2020;20(10):615‐632. 10.1038/s41577-020-00434-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Harvey WT, Carabelli AM, Jackson B, et al. SARS‐CoV‐2 variants, spike mutations and immune escape. Nat Rev Microbiol. 2021;19(7):409‐424. 10.1038/s41579-021-00573-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hadfield J, Megill C, Bell SM, et al. Nextstrain: real‐time tracking of pathogen evolution. Bioinformatics. 2018;34(23):4121‐4123. 10.1093/BIOINFORMATICS/BTY407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brasil. Boletim Epidemiológico Especial 100‐ Doença pelo Coronavírus COVID‐19/Ministério da Saúde e Secretaria de Vigilância em Saúde. February 2022. https://covid.saude.gov.br
- 8. Faria NR, Mellan TA, Whittaker C, et al. Genomics and epidemiology of P.1 SAS‐CoV‐2 lineage in Manaus, Brazil. Science. 2021;372(6544):815‐821. 10.1126/science.abh2644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Farfour E, Lesprit P, Visseaux B, et al. The Allplex 2019‐nCoV (Seegene) assay: which performances are for SARS‐CoV‐2 infection diagnosis? Eur J Clin Microbiol Infect Dis. 2020;39(10):1997‐2000. 10.1007/S10096-020-03930-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Queiroz JAS, Rampazzo RCP, Filho EBS, et al. Development of a quantitative one‐step multiplex RT‐qPCR assay for the detection of SARS‐CoV‐2 in a biological matrix. Int J Infect Dis. 2021;104:373‐378. 10.1016/J.IJID.2021.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Resende PC, Naveca FG, Lins RD, et al. COVID‐19 in Amazonas, Brazil, was driven by the persistence of endemic lineages and P.1 emergence. Nature Med. 2021;27(7):1230‐1238. 10.1038/s41591-021-01378-7 [DOI] [PubMed] [Google Scholar]
- 12. Fuentes A, Serrano‐Conde E, Roldan C, et al. Antibody response in patients admitted to the hospital with suspected SARS‐CoV‐2 infection: results from a multicenter study across Spain. Eur J Clin Microbiol Infect Dis. 2021;40(6):1343‐1349. 10.1007/S10096-020-04139-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dolscheid‐Pommerich R, Bartok E, Renn M, et al. Correlation between a quantitative anti‐SARS‐CoV‐2 IgG ELISA and neutralization activity. J Med Virol. 2022;94(1):388‐392. 10.1002/JMV.27287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fundação Oswaldo Cruz . Dashboard–Genomahcov–Fiocruz. Published 2021. Accessed September 19, 2021. http://www.genomahcov.fiocruz.br/dashboard/
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All SARS‐CoV‐2 genomes generated and analyzed in this study are available at the EpiCoV database in GISAID (https://www.gisaid.org) with accession IDs EPI_ISL_10115640, EPI_ISL_10115641, and EPI_ISL_10115642.


