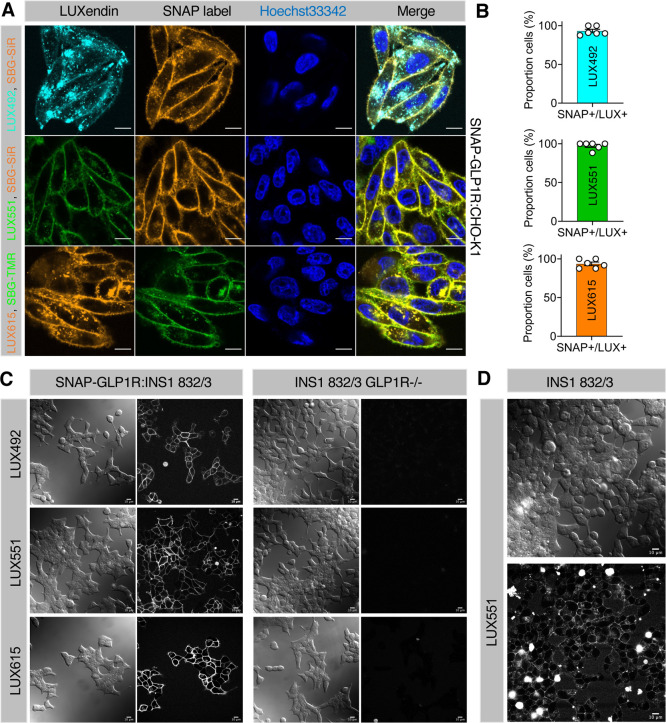
Figure 2.
(A) Labeling of live cells with LUXendin492, LUXendin551, and LUXendin615. SNAP-GLP1R:CHO-K1 cells were incubated with LUXendin492 (LUX492), LUXendin551 (LUX551), and LUXendin615 (LUX615) before orthogonal SNAP-labeling with either cell-impermeable SBG-TMR or SBG-SiR and confocal imaging (nuclei were stained using Hoechst33342) (scale bar = 10 μm) (n = three images from experiments performed in duplicate). (B) Labeling efficiency of LUXendin492, LUXendin551, and LUXendin615 in SNAP-GLP1R:CHO-K1 cells, quantified as the proportion of SNAP+/LUX+ cells (versus total SNAP-labeled cells) (n = 6 wells). (C) LUXendin492, LUXendin551, and LUXendin615 label SNAP-GLP1R:INS1 832/3 but not INS1 832/3 GLP1R–/– cells. (D) Additionally, endogenous GLP1R can be visualized with LUXendin551 in INS1 832/3 cells, which express the receptor at lower levels than in islets (scale bars = 10 μm).