Fig. 1.
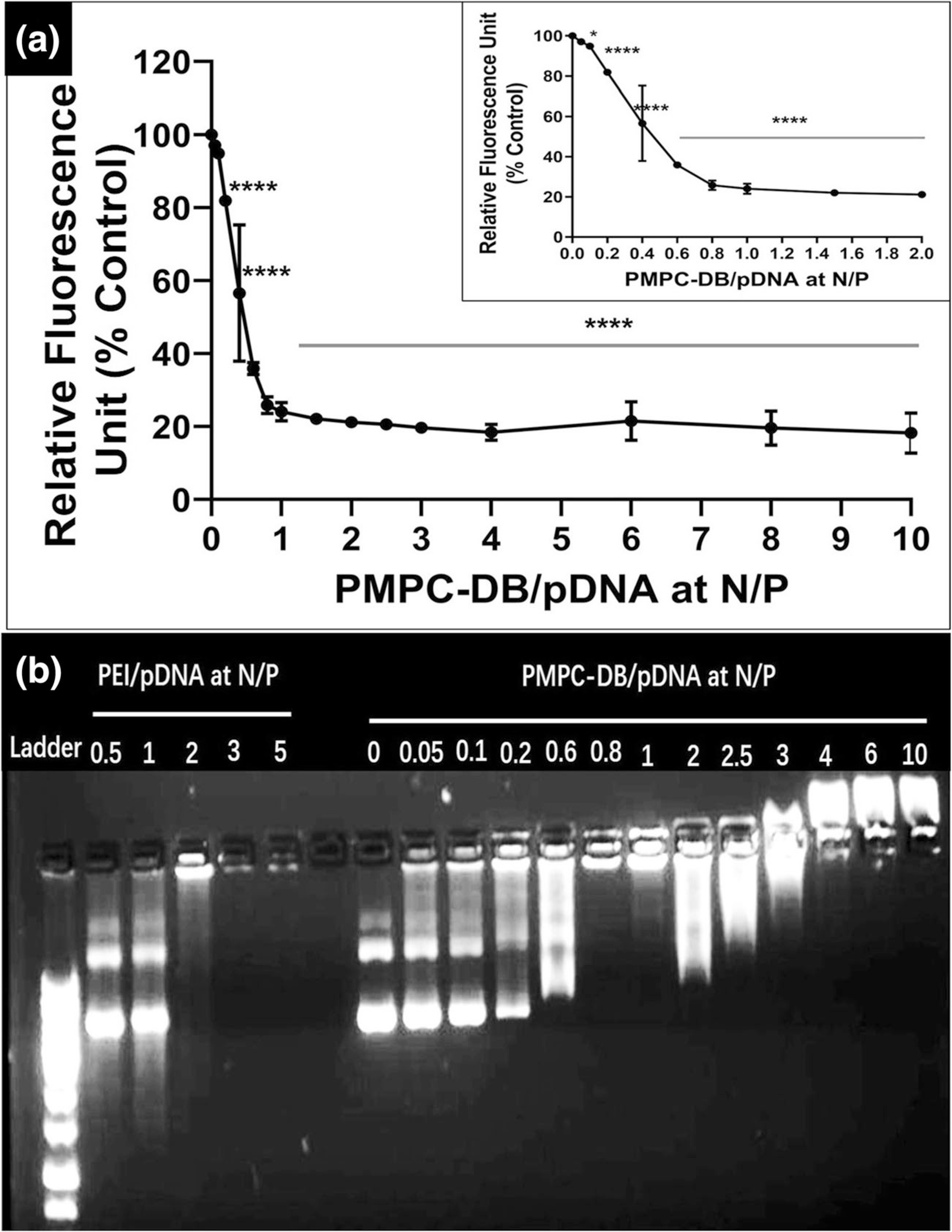
DNA condensation by PMPC-DB polymer at different N/P ratios confirmed using (a) ethidium bromide exclusion assay and (b) agarose gel electrophoresis. (a) Precalculated volumes of a PMPC-DB stock (3 mg/ml) solution was added to 20 μg/ml pDNA containing 1 μg/ml ethidium bromide in 10 mM citrate buffer pH 4.0 (N/P 0; baseline) to obtain the specified N/P ratios. The relative fluorescence unit (%) of PMPC-DB/pDNA polyplexes was calculated against the baseline. The RFU values were measured on a Fluoromax-4 spectrofluorometer. The data is presented as mean ± standard deviation (SD) from three independent experiments. One-way ANOVA was performed using GraphPad Prism 8.4.1. Asterisks indicate significant differences (****p < 0.0001, * p < 0.05). (b) Polyplexes containing 0.4 μg pDNA were loaded into a 0.8% w/v agarose gel. The gel was run at 120 V and 100 mA for 75 min. The gel was scanned under UV using a ProteinSimple red imaging system.
