Fig. 2.
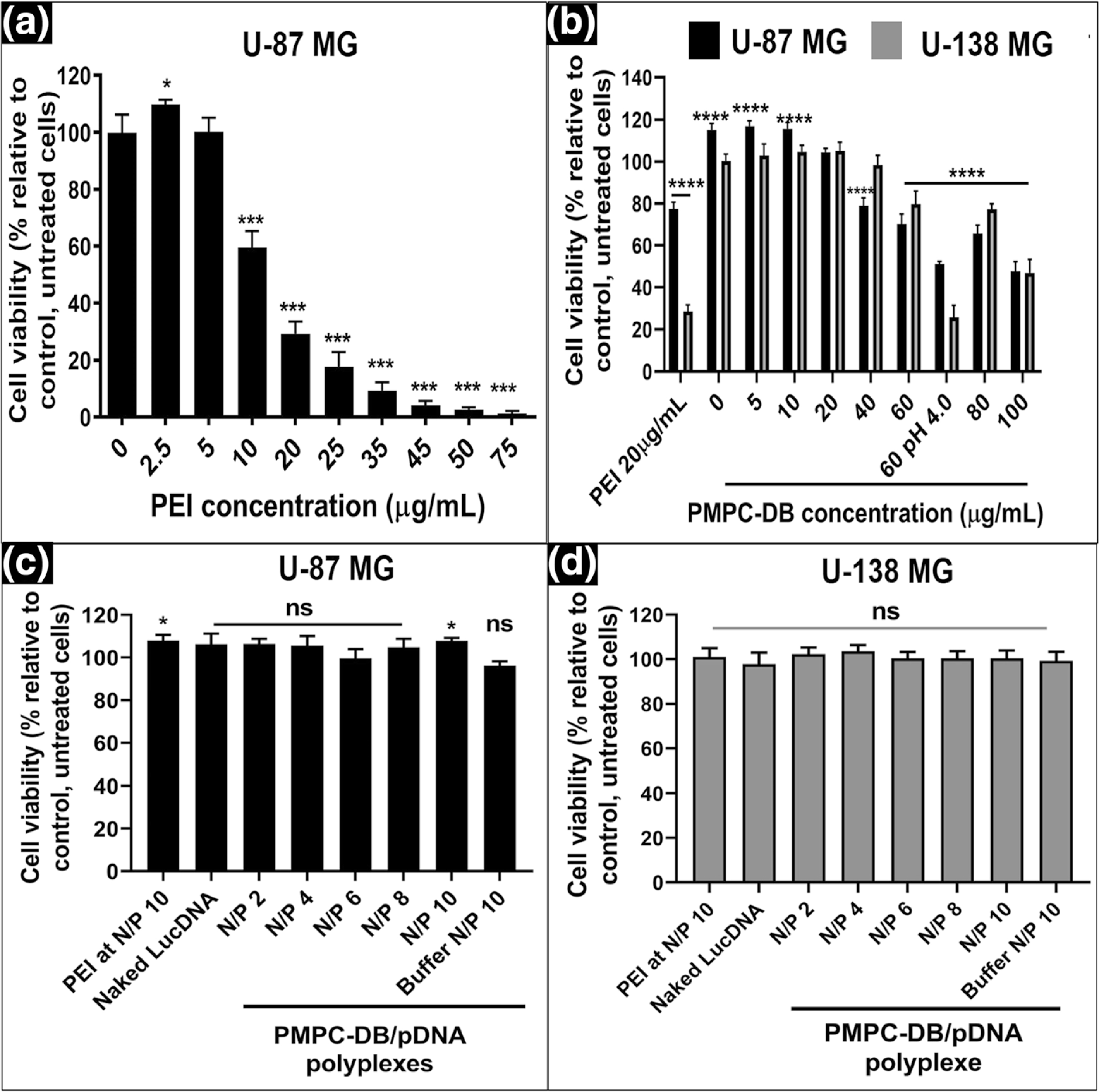
Cytocompatibility of PMPC-DB polymer and PMPC-DB/pDNA polyplexes with U-87MG and U-138MG cells using the ATP assay. U-87MG (P130) cells were seeded in 96-well plates at 16,500 cells/well. At about 80% confluency, U-87MG cells were incubated with (a) an isotonic mixture of PEI diluted in MEM/FBS medium for 4 h. U-87MG and U-138MG (P179) were (b) incubated for 4 h with PMPC-DB polymer initially dissolved in citrate buffer and 5x diluted in 10 mM phosphate buffer pH 8.0 buffer. (c-d) transfected with naked DNA, PEI/pDNA polyplexes, and PMPC-DB/pDNA at the indicated N/P ratios for 4 h. The pDNA concentration in each group was 2.8 μg/ml in a 60 μL volume/well. For ATP assays in a-d, the treatment media was replaced with fresh complete culture media and incubated for 24 h before the ATP assay. Cell Titer Glo 2.0 reagent was added at an equal volume to that of cell culture medium. The luminescence was measured at 1 s integration time using a luminometer. The % cell viability of the treated cells was calculated using the following equation: ((relative luminescence unit (RLU) of treated cells/RLU of untreated cells) × 100. The significance of treated groups was compared against control using a student’s unpaired t-test compared to control or one-way ANOVA (Bonferroni’s Multiple Comparison test) and the significance levels are indicated as *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001 (ns = non-significant)
