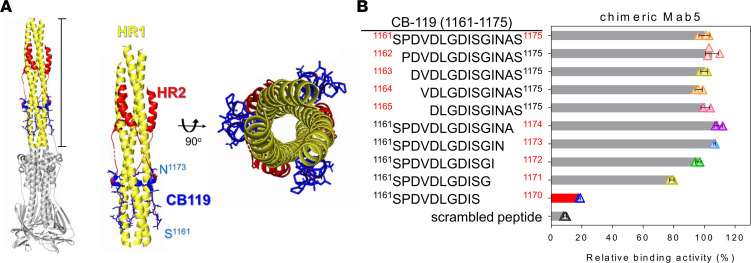
Figure 6. Structure of the fusion core and identification of the critical amino acid residues on the CB-119 epitope.
(A) Overall views of the SARS-CoV-2 S2 trimer in the postfusion machinery (left) and locations of the CB-119 epitopes (blue) in the S2 structure (Protein Data Bank: 6XRA). The zoomed-in views show the fusion core from the side view (middle), and the second view (right) has been rotated 90° to show the exposed CB-119 epitope on the surface of 3 HR1 helices. Various structural components are represented by the following color scheme: HR1 (yellow), HR2 (red), CB-119 (blue), and the N-/C-terminal residues of CB-119 (cyan). (B) Identification of the minimal Mab5 epitope was performed by ELISA. Various N- and C-terminal truncated peptides were synthesized to detect reactivity with chimeric Mab5 and labeled in the left panel with the residue numbers of the amino acids. The percentage of relative binding activity is displayed and was normalized to the binding activity level of the CB-119 peptide. The red bar indicates the weakest reaction intensity among the synthetic peptides. The data denote the means ± SDs from experiments performed independently at least 3 times.