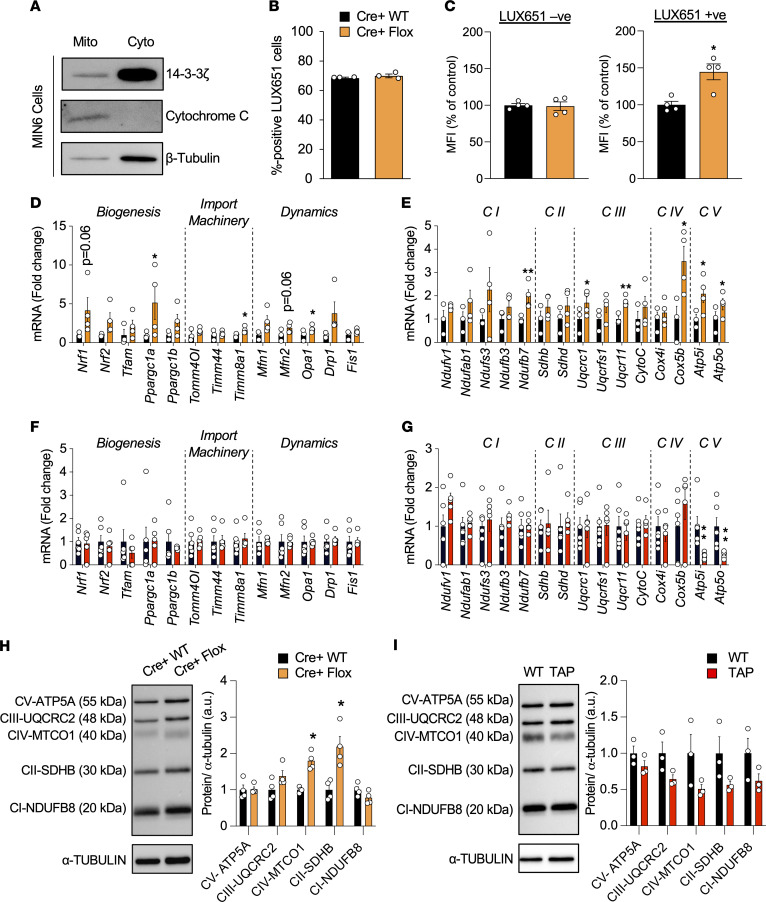
Figure 6. Detection of 14-3-3ζ in mitochondria, and analysis of its deletion, which leads to increases in mitochondrial mass and expression of genes associated with oxidative phosphorylation and biogenesis.
(A) Mitochondrial (Mito) and cytoplasmic (Cyto) fractions were obtained from MIN6 insulinoma cells, resolved by SDS-PAGE, and probed for 14-3-3ζ. Cytochrome C and β-tubulin were used as mitochondrial and cytoplasmic loading controls, respectively (n = 3 independent experiments). (B) Cre+ WT and Cre+ Flox dispersed islet preparations were incubated with LUXendin-651 (LUX651; 400 nM) for 1 hour prior to detection by flow cytometry. The proportion of LUX651+ cells (LUX+/total cells counted) represents β cells from each preparation (n = 4 per group). (C) Dispersed β14-3-3ζ–KO islets were treated by MitoTracker green (100 nM) and LUXendin-651 (400 nM) to specifically label mitochondria and β cells, respectively. Histograms depict the median fluorescence intensity (MFI) of MitoTracker green in LUX651– and LUX651+ cells (n = 4 per group; *P < 0.05 when compared with Cre+ WT). (D–G) Isolated mRNA from islets from Cre+ WT and Cre+ Flox mice (D and E) and WT and TAP mice (F and G) were subjected to qPCR analysis for mitochondrial biogenesis, import machinery, and dynamics genes (D and F), as well as for oxidative phosphorylation genes (E and G) (n = 3–4 mice per group; *P < 0.05; **P < 0.01 when compared with Cre+ WT or WT). (H and I) Western blot analysis of the OXPHOS mitochondrial complexes in islet extracts of Cre+ WT and Cre+ Flox mice (n = 3 per group; *P < 0.05 when compared with Cre+ WT mice) (H) or WT and TAP mice (n = 3 per group) (I). Significance was determined by unpaired, 2-tailed Student’s t test (C, D, E, G, and H).