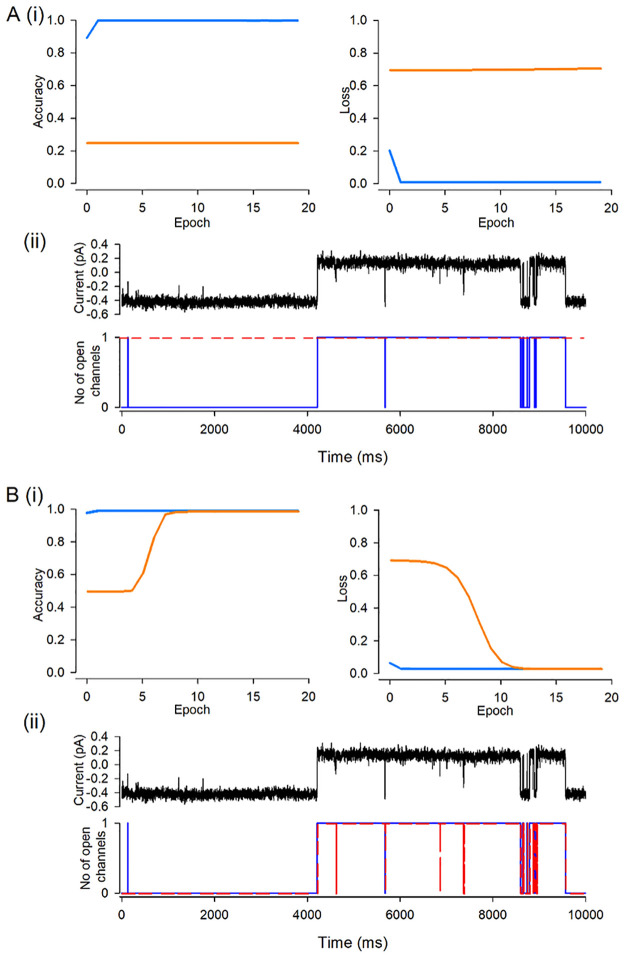
Fig 7. Potential of DeepGANnel to facilitate deep learning training on physiological time series data.
This demonstration uses data from ion channel Phenotype C. The original raw data is 150k sample points. This was split into two 75k data sets, for training and for validation by our previously published labelling network Deep-Channel [13]. (A) Shows the training and validation accuracy; the accuracy on the training set itself is near perfect, but it fails to an accuracy of 0.5 (to predict open or closed) with the unseen validation dataset. (B) Shows similar to (A), but with losses. Complete failure with the unseen validation set. Ergo, model training fails as is illustrated in (C) where the raw signal (top) is shown above the synchronised ground truth black and prediction red. In the second part of this demonstration (D, E and F), we replace the training data with the GAN generated dataset of Phenotype C, which could be any length, but in this example is 5x larger than the original. Now Deep-Channel does much better. After about 5 epochs, performance against the validation dataset improves in terms of both accuracy (D) and loss (E) and the final product is well trained model (F). There is close agreement between the ground truth (black) and the prediction (red).