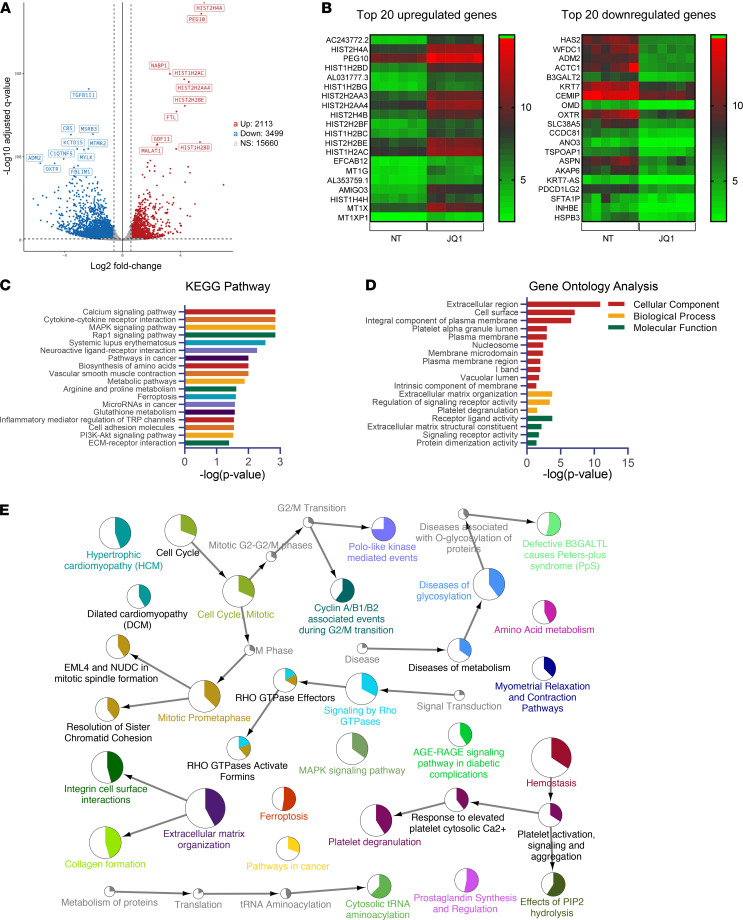
Figure 3. JQ1 treatment followed by RNA-Seq identified targets important for ECM remodeling, cell cycle regulation, and signaling in dcSSc fibroblasts.
(A) A total of 6 patient pairs were used for RNA-Seq analysis. In total, 5612 genes were significantly differentially expressed by JQ1 treatment in dcSSc fibroblasts (3499 downregulated and 2113 upregulated). (B) Top 20 upregulated and downregulated genes were shown. (C) The 18 most significantly enriched KEGG pathways after treatment with JQ1 in dcSSc fibroblasts are shown. (D) Gene Ontology analysis of the differentially expressed genes after JQ1 treatment in dcSSc fibroblasts. (E) Network of functional categories represented by JQ1-associated changes in dcSSc fibroblasts based on functional enrichment analysis. The node size corresponds with the level of significance (P value range, <0.05 to <0.0005) of the term it represents within the network. Colored portions of the pie charts represent the number of genes identified from the RNA-Seq experiment that are associated with the term. NT, no treatment. TRP, transient receptor potential.