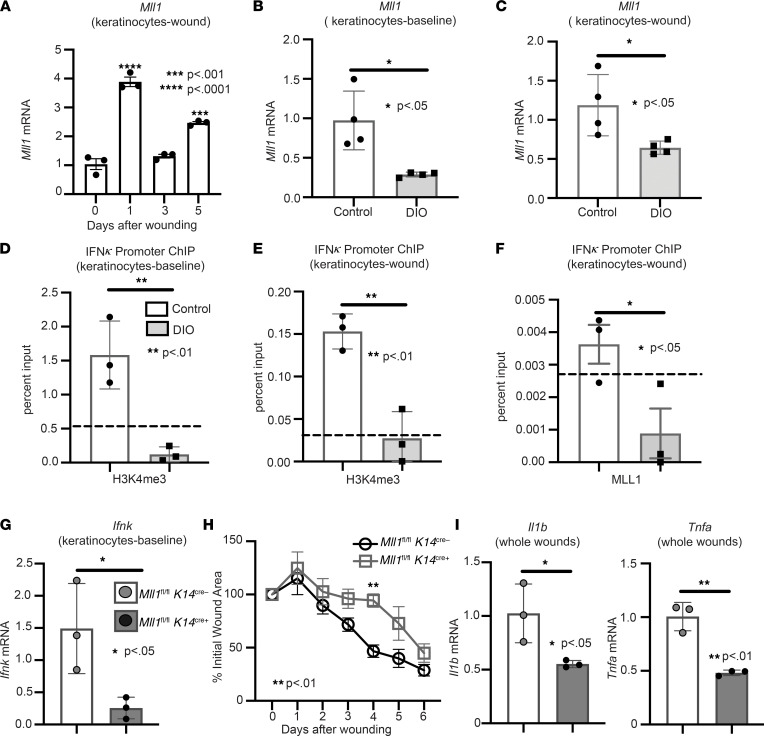
Figure 4. IFN-κ expression in diabetic keratinocytes is regulated by MLL1.
(A) Wound keratinocytes were isolated from day 0, 1, 3, and 5 wounds. Mll1 expression was examined via qPCR. n = 3 mice per group repeated 2 times in triplicate. (B and C) Keratinocytes at baseline and wounds were isolated from ND and DIO mice. Mll1 gene expression was measured via qPCR (n = 4 mice per group, repeated in triplicate). (D and E) Keratinocytes at baseline and wounds were isolated from DIO and control mice. ChIP analysis for H3K4me3 deposition at the Ifnk promoter was performed in keratinocytes from wounds and at baseline of DIO and control mice (n = 3 per group, repeated in triplicate). Respective IgG controls (dotted line). (F) Keratinocytes were isolated from DIO and control wounds. ChIP analysis for MLL1 deposition at the Ifnk promoter was performed (n = 3 per group, repeated in triplicate). The dotted line indicates the respective IgG controls. (G) Keratinocytes were isolated from Mll1fl/fl K14cre+ and littermate controls and analyzed for Ifnk expression by qPCR (n = 3 mice per group, repeated in triplicate). (H) The 4 mm punch biopsy wounds were created on Mll1fl/fl K14cre+ and littermate controls. The change in wound area was analyzed with ImageJ software (2 wounds per mouse, n = 4–5 mice per group). (I) Wounds isolated from Mll1fl/fl K14cre+ and control mice on day 3; n = 3 per group. Gene expression of inflammatory cytokines Tnf and Il1b were measured via qPCR. Data were analyzed for variances, and 2-tailed Student’s t test or 1-way ANOVA was performed. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. Data are presented as mean ± SEM.