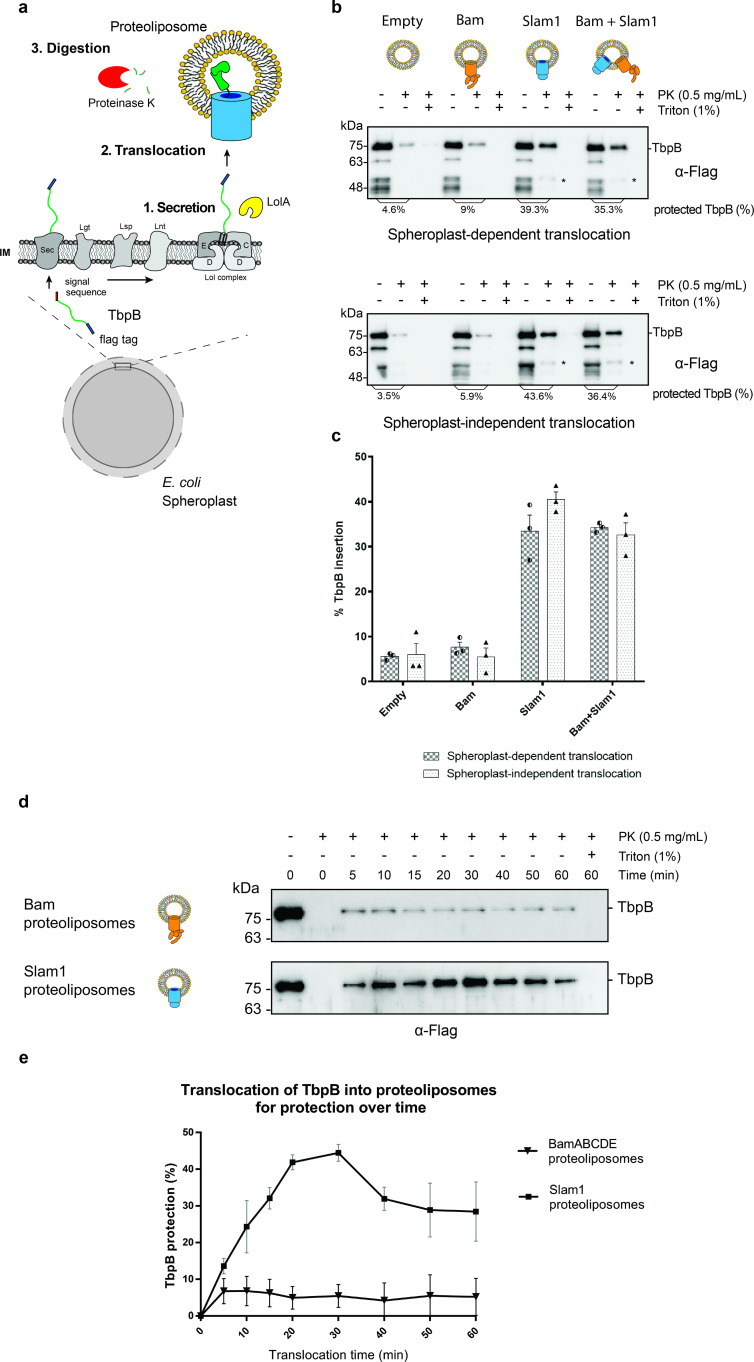
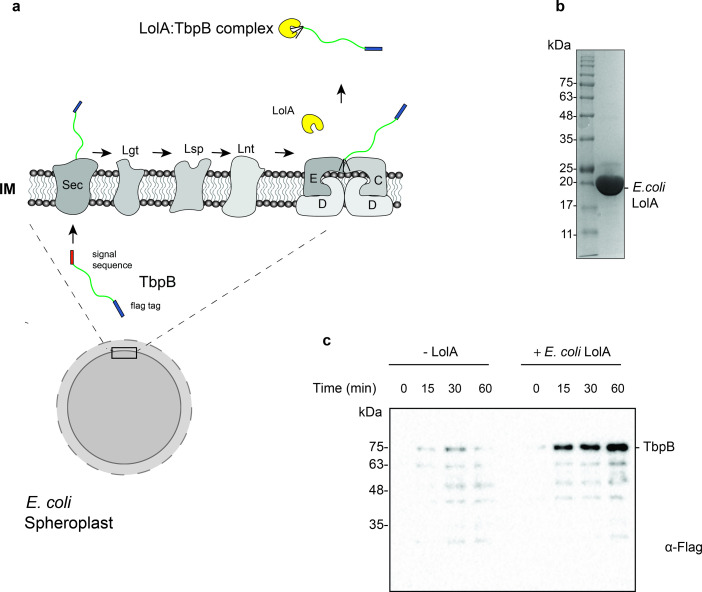
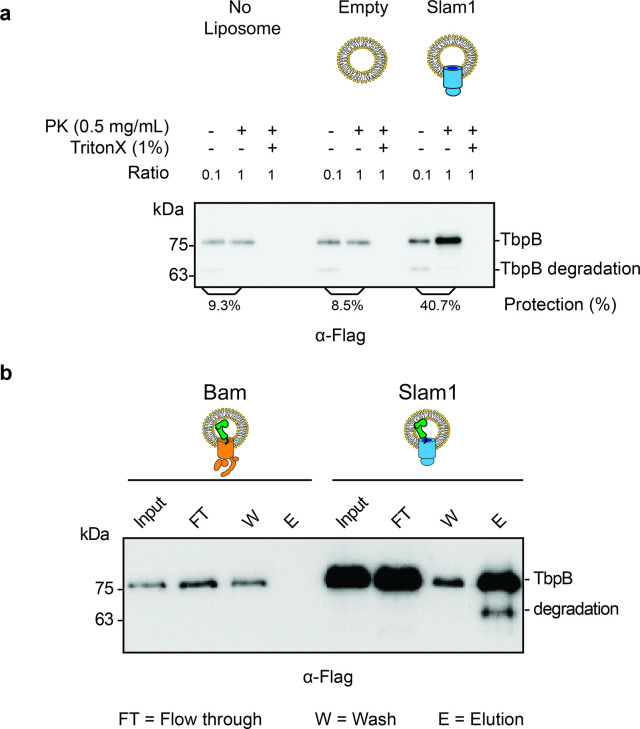
Figure 2. In vitro translocation assay for reconstitution of Slam-dependent SLP translocation.
(a) Model of the proposed in vitro proteoliposomes translocation assay for TbpB secreted directly from E. coli spheroplast. As E. coli cell expressing TbpB, the cells were converted into spheroplast that has an intact inner membrane. Purified LolA was added to release mature TbpB (processed by Lgt, Lsp, and Lnt) from the LolCDE complex in the inner membrane. The secreted TbpB was incubated with proteoliposomes for translocation, followed by PK digestion to quantify the amount of TbpB that had been translocated inside the liposomes. (b) Representative α-flag western blots obtained for the in vitro translocation assay. Slam1 proteoliposomes were incubated either with spheroplasts expressing TbpB (spheroplast-dependent translocation, upper panel) or supernatant of spheroplasts that have been induced for TbpB production (spheroplast-independent translocation, lower panel). Empty liposomes and Bam proteoliposomes were used as controls. Proteoliposomes containing Bam + Slam1 were used to test if the Bam complex plays an accessory role to Slam in TbpB translocation. For each proteoliposome, no proteinase K treatment (−PK), proteinase K treatment (+PK) and proteinase K + Triton X-100 treatment (+PK + T) samples are shown. The % TbpB protection shown was calculated by dividing the intensity of the mature TbpB band (~75 kDa) for each sample by the −PK sample. (*) Partial TbpB fragment which is only seen in the presence of Slam1 proteoliposomes. (c) Quantification of TbpB protection in proteoliposomes through densitometry analysis. The plot represents data obtained from at least three biological replicates for both spheroplast-dependent translocation and spheroplast-independent assay. Individual data points were included on the graph. (d) Representative α-flag western blot of spheroplast-independent TbpB translocation into Bam and Slam1 proteoliposomes over time. Spheroplast-secreted TbpB was incubated with proteoliposomes in 1:1 ratio at room temperature. Samples were collected every 5 or 10 min and left on ice before proteinase K treatment. (e) Quantification of spheroplast-secreted TbpB translocation into Bam proteoliposomes and Slam1 proteoliposomes over the course of 60 min.