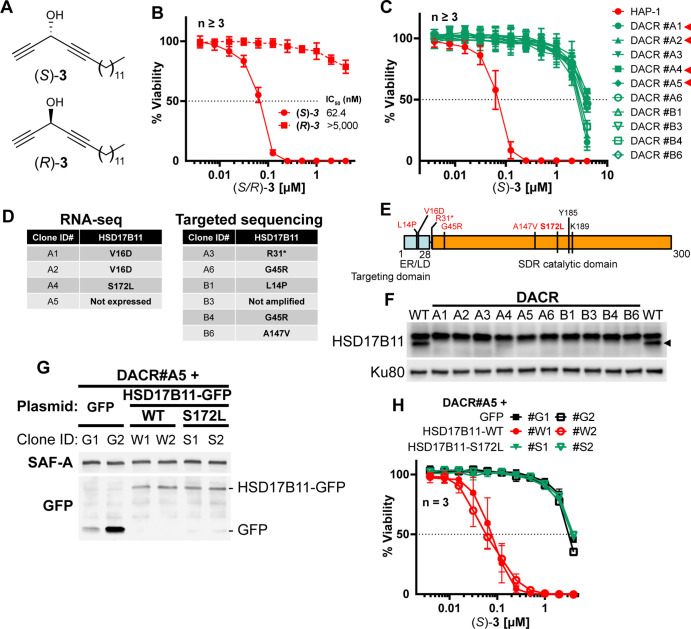
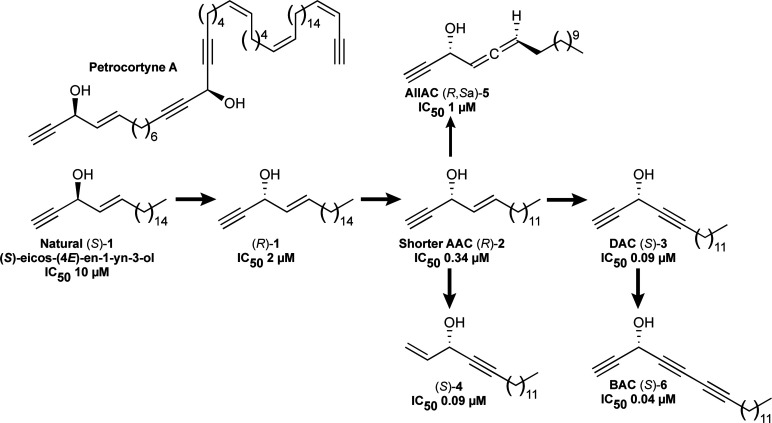
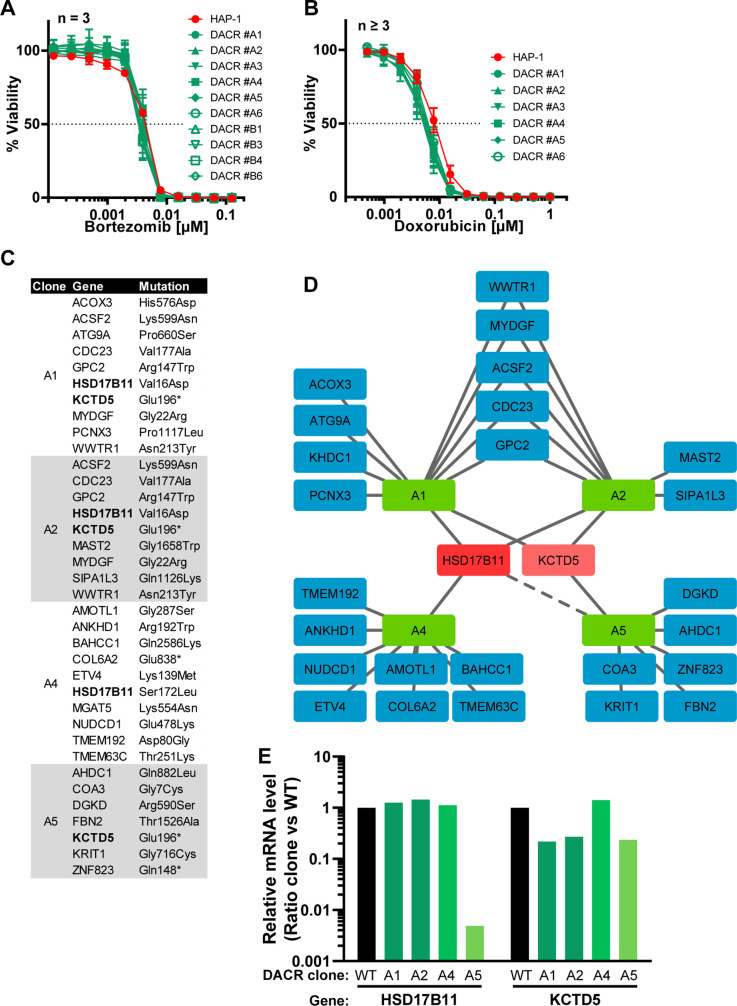
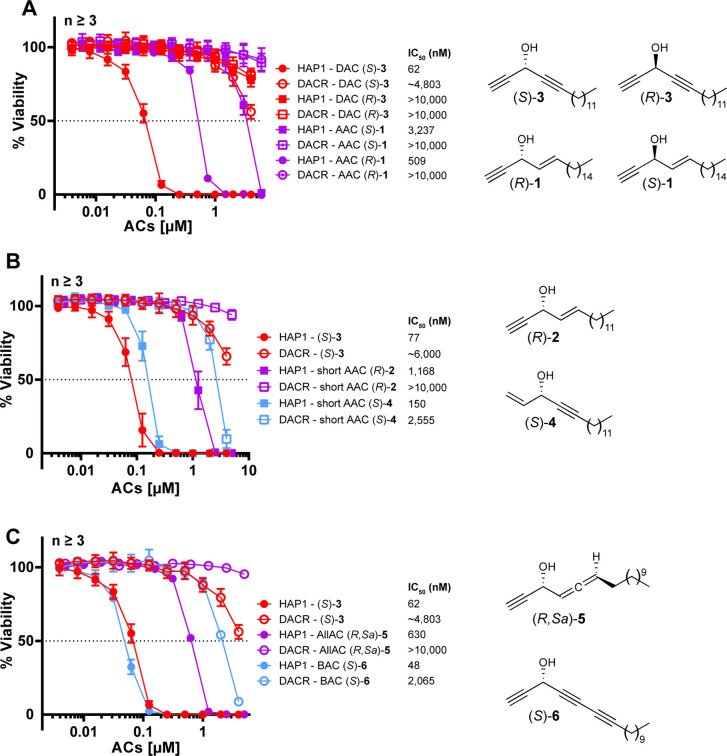
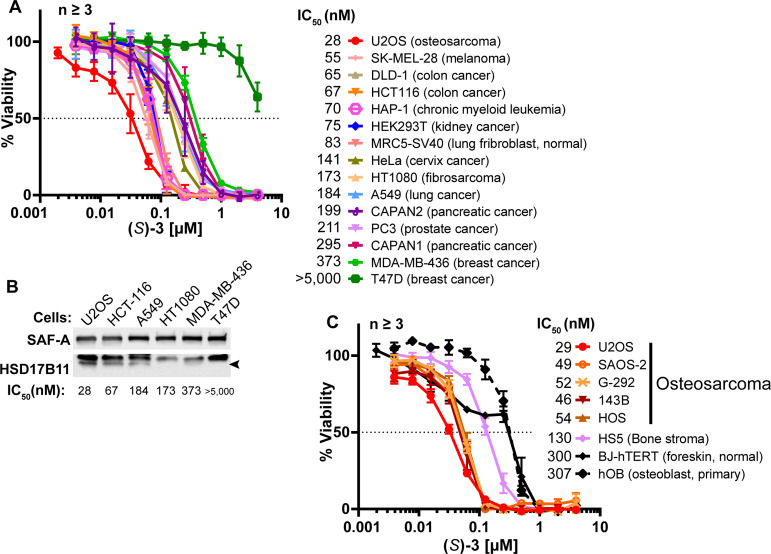
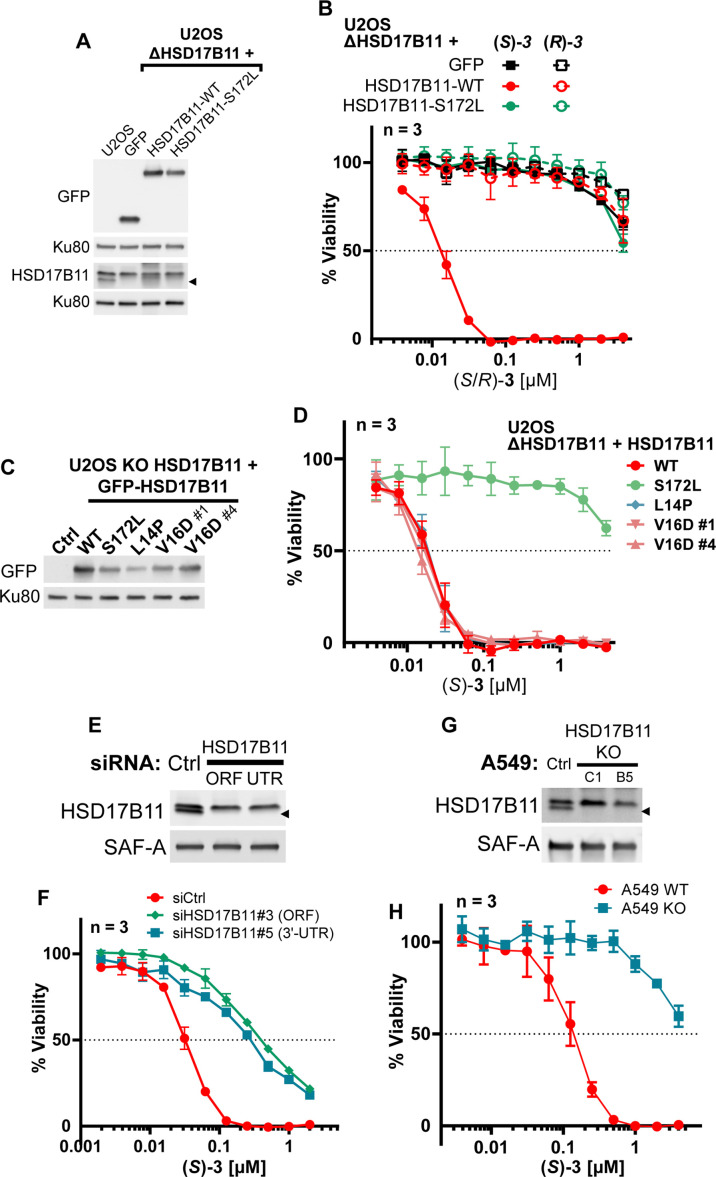
Figure 1. HSD17B11 is necessary for DAC (S)–3 cytotoxic activity.
(A) DAC (S)–3 and (R)–3 structures. (B) Cell viability analysis of HAP-1 or U2OS cells treated for 72 h with the indicated concentrations of (S)- or (R)–3. (C) Cell viability analysis of individual DAC-resistant clones or wild-type HAP-1 treated for 72 hr with the indicated concentrations of (S)–3. (D) List of mutations identified by RNA-seq or targeted sequencing of HSD17B11 in individual DAC-resistant clones. (E) Schematic representation of HSD17B11 functional domains. The positions of the identified mutations are indicated in red. The Y185, K189 (indicated in black), and S172 amino acids are critical for catalysis. (F) Analysis by immunoblotting of HSD17B11 levels in wild-type HAP-1 and DAC-resistant clones. Ku80 was used as a loading control. The black arrow indicates HSD17B11 position. (G) Analysis by immunoblotting of HSD17B11-GFP levels in individual clones of DAC-resistant clone A5 complemented with GFP, wild-type or S172L mutant HSD17B11-GFP. SAF-A and total H2AX were used as loading controls. (H) Cell viability analysis of individual clones of DAC-resistant clone A5 complemented with GFP, wild-type or S172L mutant HSD17B11-GFP treated for 72 h with the indicated concentrations of (S)–3.