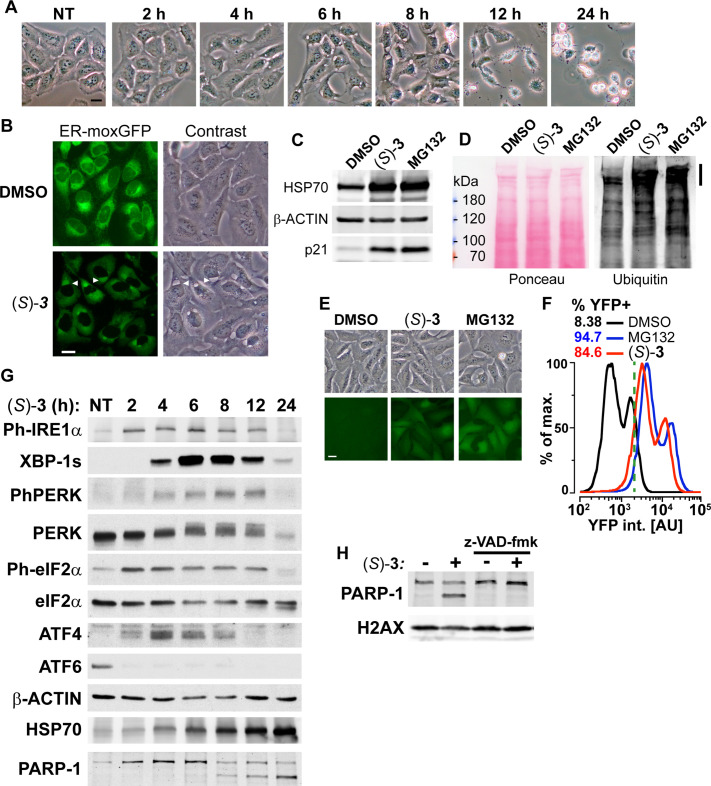
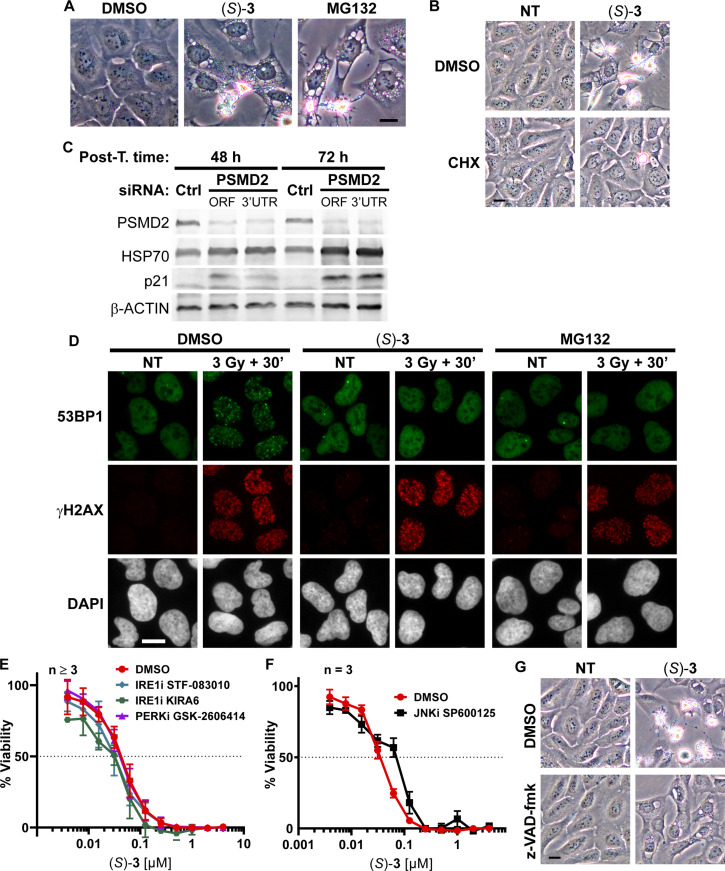
Figure 4. DAC (S)–3 triggers ER-stress, ubiquitin-proteasome system inhibition and apoptosis.
(A) U2OS stably co-expressing a GFP variant addressed and retained in the endoplasmic reticulum were treated with 1 µM (S)–3 and monitored by live imaging. Representative pictures of U2OS cells, either untreated (NT) or treated with 1 µM (S)–3 for the indicated time. (B) Representative pictures of U2OS with stably GFP-labeled endoplasmic reticulum and either untreated or treated for 8 h with 1 µM (S)–3. (C) Immunoblotting of extracts from U2OS untreated or treated with 1 µM (S)–3 or 20 µM MG132 for 8 h. (D) Immunoblotting of ubiquitin in extracts from U2OS untreated or treated with 1 µM (S)–3 or 20 µM MG132 for 2 h. High-molecular-weight ubiquitin conjugates are indicated by a vertical bar on the right. (E) Representative pictures of U2OS stably expressing Ub-G76V-YFP and either untreated or treated for 4 h with 1 µM (S)–3 or 20 µM MG132. (F) Analysis of YFP fluorescence by flow cytometry of U2OS Ub-G76V-YFP treated as described in (E) ,% of cells scored as positives using the vertical green bar as a threshold are indicated. (G) Immunoblotting using extracts from U2OS cells treated with 1 µM (S)–3 for increasing times, indicated in hours. To probe for all the UPR markers, the same extracts were analyzed on different immunoblots, each one with its loading control (see the source data), which were grouped in logical order on the figure to facilitate the interpretation. (H) Immunoblotting using extracts from U2OS cells treated with 1 µM (S)–3 for 12 h with or without 50 µM z-VAD-fmk.