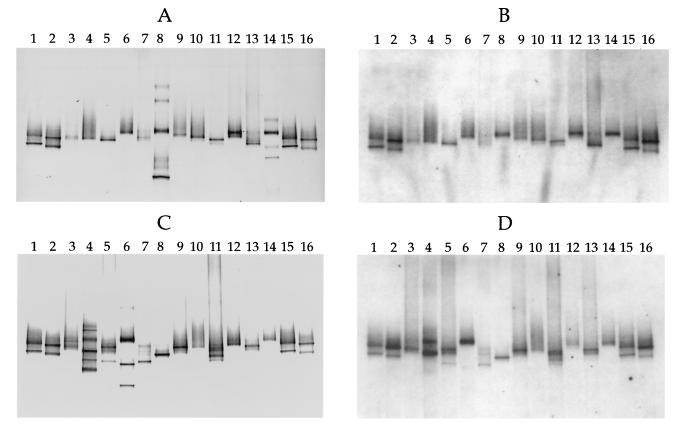
FIG. 3.
Hybridization analysis of DGGE profiles of 16S rDNA fragments obtained with primers specific for the domain Bacteria and template DNA from MPN cultures and Thiomicrospira isolates from coastal sediments of the Wadden Sea. (A) DGGE patterns. Lanes 1 and 15, T. pelophila (lower band) and Thiomicrospira sp. strain JB-B2 (upper band); lanes 2 and 16, T. kuenenii JB-A1 (lower band) and T. frisia JB-A2 (upper band); lane 3, lowest-dilution MPN culture of the sediment layer at a depth of 0 to 2 mm; lane 4, highest-dilution MPN culture of the sediment layer at a depth of 0 to 2 mm in which Thiomicrospira spp. could still be detected; lanes 5 and 6, equivalent MPN cultures from a depth of 2 to 4 mm; lanes 7 and 8, equivalent MPN cultures from a depth of 4 to 6 mm; lanes 9 and 10, equivalent MPN cultures from a depth of 6 to 8 mm; lanes 11 and 12, equivalent MPN cultures from a depth of 10 to 12 mm; lanes 13 and 14, equivalent MPN cultures from a depth of 14 to 16 mm. (B) Hybridization analysis of the DGGE pattern in panel A performed with the Thiomicrospira-specific, digoxigenin-labeled oligonucleotide, whose target sequence is located within the rDNA amplified. (C) DGGE patterns. Lanes 1, 2, 15, and 16, Thiomicrospira standards (see the explanation above for panel A); lanes 3 and 4, equivalent MPN cultures from a depth of 18 to 20 mm; lanes 5 and 6, equivalent MPN cultures from a depth of 22 to 24 mm; lanes 7 and 8, equivalent MPN cultures from a depth of 26 to 28 mm; lanes 9 and 10, equivalent MPN cultures from a depth of 30 to 32 mm; lanes 11 and 12, equivalent MPN cultures from a depth of 34 to 36 mm; lanes 13 and 14, equivalent MPN cultures from a depth of 38 to 40 mm. (D) Hybridization analysis of the DGGE pattern in panel C. Comparison of panels A and B and panels C and D shows that chemolithoautotrophic bacteria other than Thiomicrospira spp. were present in some of the MPN cultures.