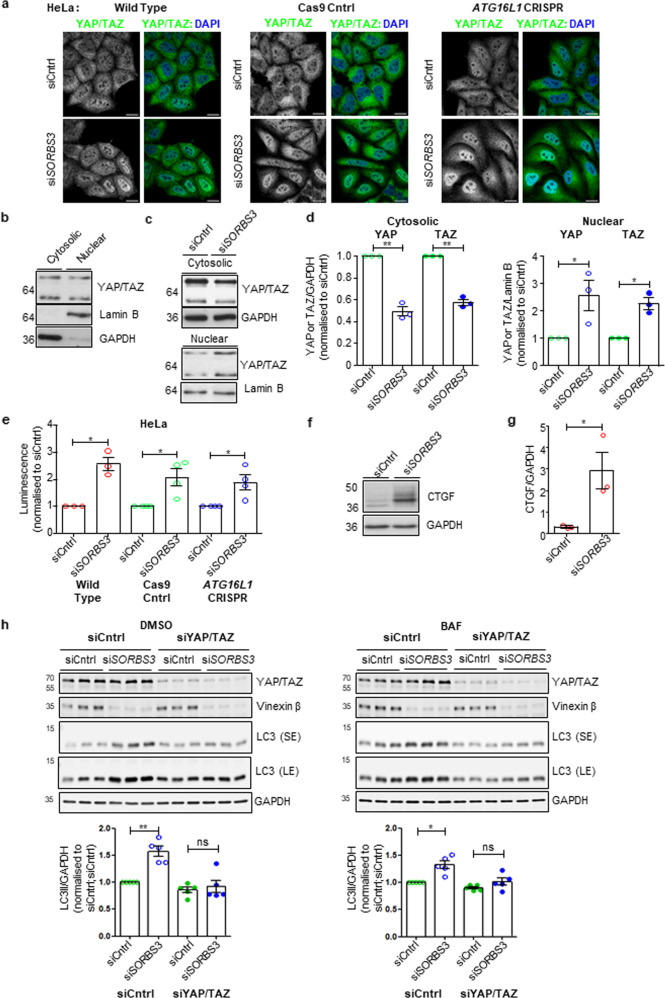
Fig. 2. Vinexin beta depletion upregulates autophagy through YAP/TAZ.
a Wild type, Cas9 control (Cntrl) and ATG16L1 CRISPR HeLa cells were depleted of vinexin beta using an individual siRNA oligonucleotide against SORBS3 (siSORBS3; oligo 7). Endogenous YAP/TAZ were examined by immunofluorescence and confocal microscopy. Representative images from three independent experiments per cell line are shown. Green = YAP/TAZ (Alexa Fluor 488); blue = DAPI. Scale bars indicate 20 µm. b Wild type HeLa cell lysate was subject to nuclear/cytosolic fractionation. Endogenous YAP/TAZ, lamin B and GAPDH protein levels in the nuclear and cytosolic fractions were examined by western blotting. Representative blot from the two independent experiments is shown. Molecular weights shown in kDa. c HeLa cells were depleted of vinexin beta using siSORBS3 (oligo 7) and lysates subject to nuclear/cytosolic fractionation and western blotting, as described in b. Representative blots from the two independent experiments in technical triplicate are shown. Molecular weights shown in kDa. d Quantification of the representative nuclear/cytosolic fractionation experiment in technical triplicate. YAP (upper band of YAP/TAZ doublet) and TAZ (lower band of YAP/TAZ doublet) are expressed relative to GAPDH (cytosolic fraction) or lamin B (nuclear fraction) loading control. YAP or TAZ/GAPDH or lamin B was normalised to control siRNA (siCntrl) treated cells. ns = p > 0.05, * = p < 0.05; ** = p < 0.01, ** = p < 0.001 by two-tailed paired t-test. Error bars indicate SEM. e Wild type, Cas9 control (Cntrl) and ATG16L1 CRISPR HeLa cells were depleted of vinexin beta, as described in a. Cells were co-transfected with synthetic TEAD (YAP/TAZ-responsive) promoter driving luciferase expression (pGL3b-8xGTIIC-luciferase) and Renilla luciferase control reporter for 24 h. Luminescence (firefly luciferase activity relative to Renilla luciferase activity) was measured by dual-luciferase reporter assay and normalised to control siRNA (siCntrl) treated cells. Quantification of 3 (wild type) or 4 (Cas9 Cntrl and ATG16L1 CRISPR) independent experiments is shown. * = p < 0.05 by two-tailed paired t-test. Error bars indicate SEM. f Wild type HeLa cells were depleted of vinexin beta, as described in a. Levels of CTGF protein (YAP/TAZ/TEAD direct target) and GAPDH (loading control) were examined by western blotting. Representative blot from the one experiment in biological triplicate is shown. Molecular weights are shown in kDa. g Quantification of 3 independent experiments as shown in f. * = p < 0.05 by two-tailed Student’s t-test. Error bars indicated SD. h (top) HeLa cells were depleted of vinexin beta using an individual siRNA oligonucleotide against SORBS3 (siSORBS3; oligo 7) and YAP/TAZ using a pools of four siRNA oligonucleotides against YAP and TAZ (siYAP/TAZ). Cells were incubated with bafilomycin A1 (BAF; 400 nM) or DMSO vehicle control for 4 h. Endogenous YAP/TAZ, GAPDH, vinexin beta and LC3 and protein levels were examined by western blotting. Representative blots from five independent experiments are shown. SE = short exposure; LE = long exposure; molecular weights shown in kDa. (bottom) Quantification of five independent experiments described in h. LC3-II (lower band of LC3 doublet) levels are expressed relative to GAPDH loading control and normalised to LC3-II/GAPDH in siCntrl treated cells. ns = p > 0.05, * = p < 0.05; ** = p < 0.01 by two-tailed paired t-test. Error bars indicate SEM.