Abstract
The present study describes the synthesis of 1,2,3-triazole–quinazolinone conjugates (5a–q) from ethyl 4-oxo-3-(prop-2-ynyl)-3,4-dihydroquinazoline-2-carboxylate and phenyl azide/substituted phenyl azides employing Cu(i) catalysed Huisgen 1,3-dipolar cycloaddition. The corresponding acids (6a–q) were obtained by hydrolysis of esters (5a–q) to study the effect of these functionalities on the biological activity. All synthesized compounds were screened for in vitro anticandidal evaluation against Candia albicans, Candida glabrata and Candida tropicalis strains. The results indicated that compound 5n showed potent anticandidal activity with IC50 in the range of 8.4 to 14.6 μg mL−1. Hemolytic activity using human red blood cells (hRBCs) and cytotoxicity by MTT assay on human embryonic kidney (HEK-293) cells revealed the non-toxic nature of the selected compounds. Growth kinetic study with compound 5n showed its fungicidal nature as no significant growth of Candida cells was observed even after 24 h. Cellular ergosterol content was determined in the presence of different concentrations of 5n to measure the activity of lanosterol 14α-demethylase indirectly. The results showed significant disruption of the ergosterol biosynthetic pathway through inhibition of lanosterol 14α-demethylase activity supported by docking studies (PDB: 5v5z). Overall, this study demonstrates the anticandidal potential of 5n which can serve as the lead for further structural optimization and SAR studies.
The present study elicits the synthesis of 1,2,3-triazole–quinazolinone conjugates (5a–q) as ergosterol inhibitors for Candida infections.
1. Introduction
Among fungal infections, the incidents of Candida infection have increased substantially in recent years particularly in immune-compromised individuals suffering from tuberculosis, cancer, AIDS and in organ transplant cases, leading to high morbidity and mortality.1,2 There are over 150 species of Candida, of which more than 17 are known to cause candidiasis in humans. The most common species causing infections are C. albicans, C. glabrata, C. tropicalis, C. parapsilosis and C. krusei.3,4 Currently used antifungal drugs for the treatment of candidiasis include polyenes, azoles, echinocandins, allylamines, and flucytosine.5 Among these, azole antifungals including fluconazole, itraconazole, voriconazole, posaconazole, miconazole etc. are used in the treatment of both superficial and invasive fungal infections due to their broader antifungal spectrum and safety profile.6 These antifungal drugs exert their effects by inhibiting the enzyme lanosterol 14α-demethylase (CYP51), which plays an important role in the ergosterol biosynthetic pathway.7 The triazole ring of azole antifungals is an essential pharmacophore responsible for tight binding to CYP51 via coordination with the heme atom.8 However, broad application of these antifungals had caused severe drug resistance, which significantly reduced their clinical efficacy.9 Thus, there is still urgency for the discovery and development of next generation of triazole antifungal agents.10 For instance, albaconazole (Fig. 1), a next generation triazole antifungal exhibiting favorable antifungal spectrum, improved pharmacokinetic properties, and acceptable toxicity profiles, has been properly investigated in clinical trials.11 It has been evaluated in phase II clinical trials by Stiefel for the oral treatment of fungal infections, including toenail fungus, distal onychomycosis and subungual onychomycosis.
Fig. 1. Designing strategy for present work based on available antifungal drugs.
In our continuous efforts to design biologically active heterocycles,12–17 we hypothesized albaconazole as a model for preparation of 1,2,3-triazole derivatives of quinazolin-4(3H)-one (5a–q and 6a–q) and studied their antifungal properties. Further, single crystal X-ray study of compounds 2 and 5c has been reported. The drug likeliness properties of all the synthesized compounds were evaluated using QikProp version 5.1, Schrödinger software. The compounds found with better antifungal activity were also accessed for cytotoxicity by hemolytic assay on human RBCs as well as MTT assay on HEK-293 cells. As earlier reports suggested that azole drugs inhibit fungal growth by interference in sterol biosynthesis,18–20 we determined the effect of our lead inhibitor on ergosterol content further confirmed by molecular docking studies with lanosterol 14α-demethylase (CYP51) from C. albicans (PDB: 5v5z).
2. Results and discussion
2.1. Chemistry
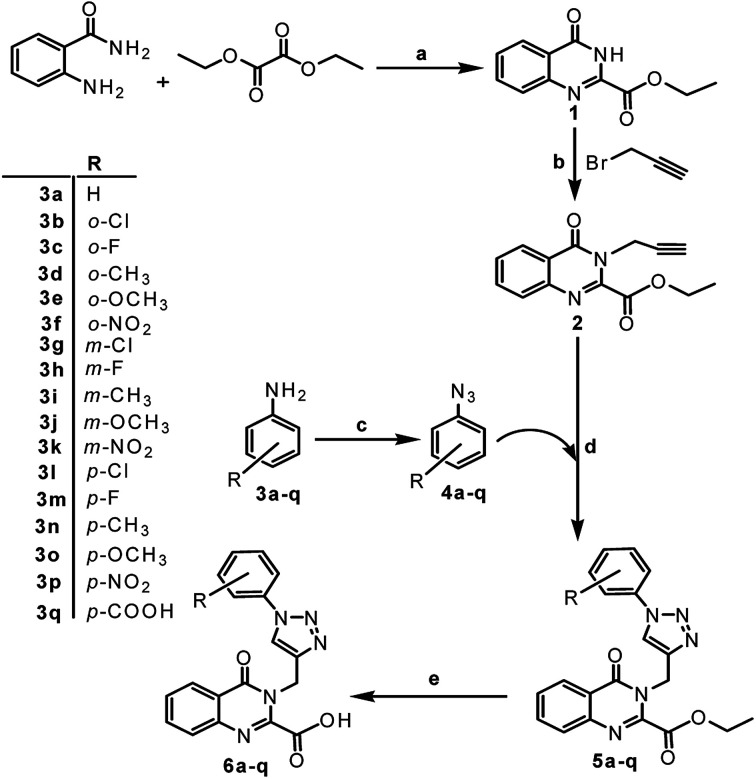
The synthetic approach exploited for the preparation of the title compounds 5a–q and 6a–q is visualized in Scheme 1. Using anthranilamide as the starting moiety, a condensation reaction was performed with diethyl oxalate at 185–186 °C for 7 h to obtain quinazolinone ester (1) in good yield.21 The coupling of 1 with propargyl bromide using K2CO3 in DMF led to the intermediate 2 containing the terminal alkyne functionality. On the other hand, aryl azides (4a–q) were prepared by the diazotization of corresponding anilines (3a–q) with NaNO2 and HCl followed by the reaction with NaN3 in a single reaction vessel. It was considered of interest to prepare various substituted phenyl azides to investigate the influence of such structural variations on the anticipated biological activities. Both the alkyne and azide components were reacted via Huisgen 1,3-dipolar cycloaddition reaction in the presence of CuSO4 and sodium ascorbate in THF : H2O (1 : 2) mixture to obtain the compounds (5a–q) in excellent yield (90–97%).
Scheme 1. Synthesis of 1,2,3-tiazole–quinazolinone conjugates. Reagents and conditions: (a) reflux (185–186 °C),5 h, 82%; (b) K2CO3, DMF, 0 °C r.t., 3 h, 92%; (c) NaNO2 in HCl, NaN3,0 °C r.t., 2.5 h; (d) CuSO4·5H2O, sodium ascorbate, THF : H2O (1 : 2), r.t., 4–5 h, 90–97%; (e) LiOH·H2O, THF : H2O (3 : 1), r.t., 2–3 h, 55–89%.
Compounds 5a–q upon hydrolysis with lithium hydroxide monohydrate (3.0 eq.) in THF : H2O mixture (3 : 1) for 2–3 h at room temperature yields the corresponding acids (6a–q) in moderate to good yield (55–89%). The hydrolysis was carried to study the effect of acid functionality on subsequent biological activity.
The structures of the newly synthesized compounds were confirmed by spectral data (1H, 13C NMR, IR, LC-MS) and elemental (CHN) analysis. All the analytical and spectral data of the synthesized compounds were in full agreement with the proposed structures. From the IR spectrum, the appearance of sharp absorption bands in the region 3105–3030 cm−1, 1743–1728 cm−1 and 1695–1654 cm−1 are ascribed to CH (triazole), C O (ester) and C O (amide) stretching frequencies, respectively. The appearance of sharp absorption bands in the region 1721–1700 cm−1 are ascribed to C O (acid) stretching frequency for compounds 6a–q, in which absorption band due to ester gets vanished. From the 1H NMR spectrum, the signals in the region at δ 8.70–6.96 ppm are attributed to aromatic protons (Ar–H), the presence of singlet at δ 8.61–7.57 ppm are attributed to CH (triazoles). The chemical shift for methylene protons (–CH2–) linking triazole ring to quinazolinone ring appears as singlet in the region δ 5.72–5.27 ppm, quartet for two H's in the region at δ 4.61–4.35 ppm belongs to –OCH2 (ester) and a triplet in the region at δ 1.49–1.19 ppm corresponds to terminal –CH3 of ester group. The signals of ester group vanish in compounds 6a–q, and broad signal in the region at δ 13.24–12.65 ppm in some of the compounds, are attributed to –OH protons. In most of these compounds this peak is not observed probably due to moist solvent. In addition, all other protons appeared at the expected chemical shifts and integral values. From the 13C NMR, the presence of signals in the region at δ 161.59–160.00 ppm corresponds to (C O) amide, signals in the region at δ 161.39–161.09 ppm corresponds to (C O) ester and in case of acids signals in the region at δ 166.81–161.04 ppm corresponds to (C O) acids. The carbon signals of –OCH2 (ester) appear in the range 63.83–63.39 ppm and terminal –CH3 of the ester group in the range δ 14.02–13.93 ppm. The signals of methylene (–CH2–) linking triazole ring to quinazolinone ring appear in the range δ 41.81–39.13 ppm while all other carbons show peaks at their expected values. The mass spectra of the compounds possessing ester group (5a–q) exhibited either [M + H]+ or [M − H]− and those possessing carboxylic acid group (6a–q) exhibited [M + H]+, [M − H]− or [M–COOH]+ which further confirmed the formation of desired compounds.
2.2. X-ray crystallographic analysis
X-ray crystallographic determination of compounds 2 and 5c was performed to support the structural analysis data. A colorless crystal of compound 2 (C14H12N2O3) and 5c (C20H16N5O3F) with approximate dimensions 0.5 mm × 0.05 mm × 0.05 mm and 0.7 mm × 0.5 mm × 0.2 mm respectively were used for the X-ray crystallographic analysis. Crystal structure analysis of compound 2 and 5c reveals that they crystallize in monoclinic crystal system with space group P21/n and P21/c, respectively. The data collection and structure refinement details are described in ESI (Table S10†). For compound 2, 19 112 reflections collected were collected out of which 2591 reflections were independent with Rint = 0.0522 and for compound 5c, 11 976 reflections collected were collected out of which 3629 reflections were independent with Rint = 0.0372. Finally, after refinement, a reasonably good R factor was found (R = 0.0427) and (R = 0.039) for compounds 2 and 5c respectively. The crystal structure of compounds is given in Fig. 2.
Fig. 2. Single crystal X-ray diffraction structures of compounds (a) 2; and (b) 5c. Structures have drawn using Olex2 with ellipsoids at 50% probability.
2.3. Pharmacological evaluation
2.3.1. Anticandidal evaluation
The inhibitory potential of synthesized 1,2,3-triazole–quinazolinone conjugates (5a–q) and (6a–q) was evaluated against three different strains of Candida: C. albicans (ATCC 90028), C. glabrata (ATCC 90030) and C. tropicalis (ATCC 750) and the results were compared with the standard drug fluconazole (FLC). The IC50 values were estimated in μg mL−1 and given in Tables 1 and 2. The in vitro results showed that all the compounds possess anticandidal activity to a certain extent.
In vitro anticandidal activity of compounds (5a–q) against Candida spp.

| ||||
|---|---|---|---|---|
| Comp. | R | IC50 ± S.D (μg mL−1) | ||
| C. albicans | C. glabrata | C. tropicalis | ||
| 5a | H | 227.7 ± 3.8 | 181.8 ± 7.5 | 74.7 ± 2.1 |
| 5b | o-Cl | 124.4 ± 18.4 | 183.4 ± 12.3 | 65.6 ± 4.3 |
| 5c | o-F | 109.9 ± 4.8 | 244.7 ± 17.5 | 87.1 ± 2.2 |
| 5d | o-CH3 | 297.2 ± 5.9 | 351.3 ± 6.3 | 191.0 ± 17.2 |
| 5e | o-OCH3 | 18.6 ± 3.4 | 81.8 ± 4.7 | 13.8 ± 2.5 |
| 5f | o-NO2 | 269.4 ± 5.5 | 272.4 ± 4.4 | 238.5 ± 7.1 |
| 5g | m-Cl | 65.6 ± 3.2 | 58.2 ± 2.4 | 28.2 ± 2.2 |
| 5h | m-F | 509.5 ± 6.4 | 364.8 ± 3.4 | 182.1 ± 11.6 |
| 5i | m-CH3 | 352.6 ± 11.1 | 395.1 ± 7.5 | 137.1 ± 6.5 |
| 5j | m-OCH3 | 340.5 ± 7.4 | 417.0 ± 2.3 | 113.2 ± 3.7 |
| 5k | m-NO2 | 142.6 ± 2.5 | 354.04 ± 5.4 | 117.0 ± 3.2 |
| 5l | p-Cl | 215.5 ± 3.5 | 194.9 ± 3.4 | 249.7 ± 9.7 |
| 5m | p-F | 209.3 ± 6.5 | 292.5 ± 9.1 | 434.9 ± 15.5 |
| 5n | p-CH3 | 14.6 ± 2.1 | 8.4 ± 1.3 | 12.4 ± 3.2 |
| 5o | p-OCH3 | 394.3 ± 1.7 | 118.2 ± 6.5 | 441.7 ± 4.3 |
| 5p | p-NO2 | 151.2 ± 15.7 | 315.5 ± 8.9 | 213.8 ± 12.1 |
| 5q | p-COOH | 342.3 ± 5.7 | 87.7 ± 5.6 | 290.2 ± 6.7 |
| FLCa | 15.6 ± 2.2 | 7.8 ± 1.5 | 8.5 ± 1.7 | |
FLC = fluconazole.
In vitro anticandidal activity of compounds (6a–q) against Candida spp.
| ||||
|---|---|---|---|---|
| Comp. | R | IC50 ± S.D (μg mL−1) | ||
| C. albicans | C. glabrata | C. tropicalis | ||
| 6a | H | 118.7 ± 7.6 | 566.1 ± 9.4 | 184.1 ± 4.8 |
| 6b | o-Cl | 168.8 ± 3.4 | 266.5 ± 10.3 | 210.6 ± 13.3 |
| 6c | o-F | 100.7 ± 3.7 | 174.4 ± 3.1 | 24.8 ± 2.7 |
| 6d | o-CH3 | 131.2 ± 4.3 | 357.7 ± 6.8 | 233.1 ± 6.9 |
| 6e | o-OCH3 | 48.2 ± 3.2 | 29.4 ± 4.2 | 9.1 ± 2.7 |
| 6f | o-NO2 | 700.8 ± 23.6 | 486.6 ± 18.5 | 520.7 ± 24.1 |
| 6g | m-Cl | 100.3 ± 4.0 | 278.1 ± 4.0 | 244.6 ± 2.0 |
| 6h | m-F | 134.8 ± 2.4 | 261.9 ± 13.3 | 175.2 ± 3.6 |
| 6i | m-CH3 | 133.4 ± 4.2 | 244.8 ± 11.2 | 222.8 ± 18.7 |
| 6j | m-OCH3 | 214.3 ± 11.0 | 453.3 ± 13.2 | 444.1 ± 17.4 |
| 6k | m-NO2 | 700.8 ± 23.6 | 486.6 ± 18.5 | 520.7 ± 24.1 |
| 6l | p-Cl | 220.9 ± 11.2 | 517.0 ± 18.6 | 303.3 ± 8.5 |
| 6m | p-F | 171.9 ± 5.3 | 421.6 ± 15.4 | 231.2 ± 5.7 |
| 6n | p-CH3 | 87.0 ± 4.0 | 288.3 ± 7.4 | 13.5 ± 3.0 |
| 6o | p-OCH3 | 124.9 ± 5.9 | 718.4 ± 13.2 | 162.3 ± 5.2 |
| 6p | p-NO2 | 789.2 ± 16.4 | 432.5 ± 10.3 | 336.6 ± 11.1 |
| 6q | p-COOH | 916.3 ± 15.5 | 522.0 ± 4.0 | 457.3 ± 10.7 |
| FLC | 15.6 ± 2.2 | 7.8 ± 1.5 | 8.5 ± 1.7 | |
Moreover, the compounds 5g, 5n and 6e showed better activity against all the three strains of Candida, viz. C. albicans, C. glabrata and C. tropicalis with IC50 values in the range 8.4 μg mL−1 to 65.6 μg mL−1. Interestingly, compound 5n showed the IC50 values against the tested strains of Candida similar to the IC50 values for standard drug FLC. Thus 5n emerging as the most potent inhibitor among the series. Similarly, for compound 6e the IC50 value against C. tropicalis (9.1 μg mL−1) is comparable with the IC50 values for standard drug FLC (8.5 μg mL−1). The compound 5e showed good among the series. Similarly, for compound 6e the IC50 value against C. tropicalis (9.1 μg mL−1) is comparable with the IC50 values for standard drug FLC (8.5 μg mL−1). The compound 5e showed good anticandidal activity against C. albicans and C. tropicalis with IC50 values of 18.6 μg mL−1 and 13.8 μg mL−1 respectively, whereas, moderate activity against C. glabrata with IC50 values of 81.8 μg mL−1. The compounds 5a, 5b and 5c showed moderate activity against C. tropicalis with IC50 values of 74.7 μg mL−1, 65.6 μg mL−1 and 87.1 μg mL−1, respectively. The compound 5g also showed moderate activity against the three Candida strains with IC50 values 65.6 μg mL−1, 58.2 μg mL−1 and 28. μg mL−1, respectively. Further, compound 6n showed moderate activity against C. albicans with an IC50 value of 87.0 μg mL−1 and good activity against C. tropicalis with IC50 values of 13.5 μg mL−1.
Structure-activity relationship (SAR) suggested that there is no clear activity relationship, in case of compounds (5a–q) the compounds bearing unsubstituted phenyl group (5a) and those bearing o–chlorophenyl (5b) and o–fluorophenyl group (5c) on 1,2,3-triazole ring showed moderate activity against C. tropicalis while weak activity against C. albicans and C. glabrata and those bearing o-methylphenyl (5d) and o-nitrophenyl group (5f) showed weak activity against all the three Candida strains. Whereas, compound bearing o-methoxyphenyl group (5e) showed good activity against C. albicans and C. tropicalis but weak activity against C. glabrata. Among meta-substituted compounds, only those bearing m-chlorophenyl group (5g) showed moderate to good activity against all the three Candida strains and all other meta-substituted derivates showed weak activity. Similarly, among para substituted derivatives, the compound bearing p-methylphenyl group (5n) showed good activity against all the three Candida strains and is the lead inhibitor among the synthesized compounds. All other compounds among para substituted derivatives showed weak activity except p-COOH (5q) which showed moderate activity (IC50 = 87.7 ± 5.6 μg mL−1) against C. glabrata.
In the case of corresponding acids (6a–q), the data showed no improvement in activity on hydrolysis of the ester functionality of compounds (5a–q) with few exceptions. For instance, in compound 6c, there is increase an in activity compared to 5c and in compound 6e there is an increase in activity against C. glabrata and C. tropicalis compared with compound 5e. Further, for compound 6n there is a decrease in activity compared with 5n, a lead inhibitor amongst compounds (5a–q).
2.3.2. Cytotoxicity by hemolytic assay
Based on in vitro anticandidal activity results, compounds 5e, 5g, 5n, 6c, 6e and 6n were selected to check their cytotoxic effect on human red blood cells (hRBCs) by the hemolytic assay. The toxicity of standard drug (FLC) was also determined for reference. At the concentration of 1000 μg mL−1, all compounds except 5n showed less than 20% hemolysis indicating their low toxicity. Compound 5n showed 41.20% hemolysis which is still less than that of standard drug fluconazole viz. 78.10% (Fig. 3). The results indicate that all the seven compounds have a very low toxic effect on human RBCs than the standard drug fluconazole under experimental conditions.
Fig. 3. Hemolytic activity of compounds 5e, 5g, 5n, 6c, 6e and 6n on hRBCs.
2.3.3. Cytotoxicity by MTT assay
The compounds with comparatively lower IC50 values (5e, 5g, 5n, 6c, 6e, and 6n) were screened for cytotoxicity against human embryonic kidney (HEK-293) cells and the results are shown in Fig. 4. A sub-confluent population of HEK-293 cells was treated with increasing concentration of each compound, and the number of viable cells was measured after 24 h by MTT cell viability assay. The cytotoxicity of all tested compounds was found to be concentration dependent. From the results, it is evident that even at 100 μg mL−1 concentrations of the compounds, no significant decrease in cell viability was observed, indicating their non-cytotoxic nature. Based on the in vitro anticandidal as well as cytotoxicity data, compound 5n was selected for further biological studies.
Fig. 4. Cell viability assay on HEK-293 cell line.
2.3.4. Growth curve studies
To investigate the effect of the potent compound 5n on the growth of Candida cells, growth kinetic studies were performed. The Candida cells were exposed to different concentrations of the test compound (2MIC, MIC and MIC/2). FLC treated cells were used as positive control and untreated cells were used as negative control. The Candida cells did not show any significant growth when exposed to the 2MIC and MIC concentrations of the above compound with a continuous lag phase of 24 h. However, at sub-MIC concentrations of 5n, growth was observed after 7 h, for standard C. albicans strain. The results showed fungicidal nature of compound 5n as no significant growth observed even after 24 h at higher concentrations. The study suggested that compound 5n to be a potent inhibitor of Candida cell growth (Fig. 5).
Fig. 5. Dose-dependent growth curve of C. albicans in the presence of 2MIC, MIC and MIC/2 concentrations of compound 5n.
2.3.5. Sterol estimation assay
Azole drugs are known to inhibit the growth of a fungal cell by an interruption in the sterol biosynthetic pathway. These drugs bind with lanosterol 14α-demethylase, an intermediate enzyme of sterol biosynthetic pathway, which result in the depletion of ergosterol.20 To support this argument in case of azole drugs, we also determined the ergosterol content in the presence of our lead inhibitor 5n by sterol quantitation method in C. albicans ATCC 90028.22,23 UV spectrophotometric sterol profile of C. albicans showed a dose-dependent sharp decrease in sterol content as shown in Fig. 6. If compared to untreated cells i.e. no inhibition in ergosterol biosynthesis, 57, 55 and 48% decrease in ergosterol content was observed in C. albicans ATCC 90028 at 2MIC (500 μg mL−1), MIC (250 μg mL−1) and MIC/2 (125 μg mL−1) concentration, respectively (Fig. 6b).
Fig. 6. Sterol estimation assay: (a) UV spectrophotometric sterol profiling graph showing flat line at 2MIC and MIC treated samples indicated very low concentration of sterol; (b) bar graph showing % decrease in ergosterol content.
Thus a significant decrease in ergosterol content is a clear indication of the disruption of the sterol biosynthetic pathway. Moreover, we also did molecular docking studies with lanosterol 14α-demethylase from C. albicans (PDB: 5v5z) to support these results.
2.4. Docking studies
As discussed in the previous section, azole drugs bind with lanosterol 14α-demethylase and inhibit ergosterol biosynthesis in the fungal cell. Lanosterol 14α-demethylase enzyme (CYP51) is a member of cytochrome P450 superfamily which catalyses the oxidative removal of 14α methyl group from lanosterol in ergosterol biosynthesis pathway in fungi. It is a heme-thiolate enzyme which converts lanosterol into 4,4′-dimethyl-cholesta-8,14,24-triene-3-β-ol. Inhibition of CYP51 leads to depletion of ergosterol coupled with an accumulation of 14-methyl sterols which results in impaired cell growth in fungi.
Recently, Keniya et al. successfully isolate CYP51 from pathogenic yeast C. albicans and co-crystallized the protein with known inhibitor 1YN (PDB: 5v5z).24 Here, we performed in silico molecular docking studies of 5n with CaCYP51 to validate our sterol quantitation analysis. Interestingly, the study revealed that compound 5n showed polar interaction with heme atom in the binding site. Heme plays a key role in the oxidation of lanosterol. The binding affinity and bond length of interaction was −9.8 kcal mol−1 and 3.6 Å, respectively. The binding pocket of compound 5n was compared with inhibitor 1YN which showed almost the same residues. Moreover, they both overlap to each other at same binding pocket. The binding site residues and interaction are shown in Fig. 7. Thus, docking results strongly support that azole drugs bind with 14α-demethylase (CYP51) and inhibit ergosterol biosynthesis which ultimately arrests cell growth, similarly observed in case of our lead inhibitor 5n.
Fig. 7. Docking of 5n with lanosterol 14α-demethylase of pathogen C. albicans (PDB: 5v5z): (a) compound 5n (yellow) showing interaction with Heme porphyrin ring (violet) which involves in oxidation process; (b) overlapping image of original inhibitor of protein 1YN (green) and compound 5n within the same binding pocket.
2.5. ADME properties
Absorption, distribution, metabolism and excretion (ADME) data have become prominent in the drug discovery process since most of the drugs fail at clinical trial due to their poor pharmacokinetic properties. These late-stage failures significantly contribute to the cost of new drug discovery projects. Considering the cost involved in the experiments to obtain the ADME data, the in silico approach provides a cost-effective alternative to filter and optimize the leads in the early phase of drug discovery.25 With this objective, in silico study of all the synthesized compounds (5a–q) and (6a–q) was performed for assessment of ADME properties and values obtained are depicted in ESI Tables S11 and S12.† We have analyzed various pharmaceutically relevant properties like, molecular weight (MW), solvent accessible surface area (SASA), total polar surface area (PSA), number of rotatable bonds (NRB), predicted aqueous solubility (QP log S), prediction of binding to human serum albumin (QP log Khsa), predicted brain/blood partition coefficient (QP log BB) and violation of Lipinski's rule of five (VLR5) using QikProp version 5.1, Schrödinger software. As per Lipinski's rule of five, a molecule likely to be developed as an orally active drug candidate should show no more than one violation of the following criteria: QP log Po/w (octanol–water partition coefficient) ≤ 5, molecular weight ≤ 500, number of hydrogen bond acceptors (HBA) ≤ 10 and number of hydrogen bond donors (HBD) ≤ 5.26 The prediction of in silico ADME parameters suggest that the synthesized compounds follow the criteria for orally active drug and thus represent a pharmacologically active framework indicating that 1,2,3-triazole–quinazolinone conjugates have new opportunities for the possible modification and future discovery of drug candidates in medicinal chemistry.
3. Experimental protocol
3.1. Materials and methods
All chemicals and reagents were purchased from Sigma-Aldrich, Merck, Spectrochem and Hi Media and were used without further purification. Reactions were monitored by TLC, performed on precoated Aluminium sheets (Silica gel 60 F254, Merck Germany) and spots were visualized under UV light at 254 nm. Purification of the compounds was done by column chromatography using silica gel (230–400 mesh size) with cyclohexane–ethyl acetate as eluent. The IR spectra of compounds were taken on Agilent Cary 630 FT-IR spectrometer. 1H and 13C NMR spectra were obtained in CDCl3/DMSO-d6 as a solvent with tetramethylsilane (TMS) as an internal standard on a Bruker Spectrospin DPX-300 spectrometer at 300 MHz. Chemical shifts (δ) are expressed in ppm and coupling constants (J) are expressed in Hertz (Hz). Multiplicities of NMR signals are represented as singlet (s), broad singlet (brs), doublet (d), double doublet (dd), triplet (t) and multiplet (m). Mass spectra were obtained on Agilent ion trap-6320 LC-MS spectrometer. Melting points were determined on a digital Buchi M-560 melting point apparatus and were reported uncorrected. Elemental analysis was performed on ElementarVario Analyzer.
3.2. Synthesis
3.2.1. Synthesis of ethyl 4-oxo-3,4-dihydroquinazoline-2-carboxylate (1)
A mixture of anthranilamide (5 g, 1.0 eq.) and diethyl oxalate (12 mL) was refluxed in an oil bath at 185–186 °C for 7 h. After completion of the reaction, monitored by TLC, the solution was allowed to cool to room temperature and excess diethyl oxalate removed under vacuum. Recrystallization from ethanol produced 1 in good yield (6.17 g).
White crystalline (needle shaped) solid, mp: 191–193 °C (lit, 192–194 °C) yield: 77%, Rf (1% methanol in DCM): 0.53, anal (C11H10N2O3) calc. C 60.55 H 4.62 N 12.84 found: C 60.51 H 4.58 N 12.78. 1H-NMR (300 MHz, CDCl3) (δ, ppm): 10.26 (brs, 1H, NH), 8.38 (dd, J = 9.3 Hz, 1H, CH), 7.99–7.96 (m, 1H, Ar–H), 7.91–7.83 (m, 1H, Ar–H), 7.66–7.60 (m, 1H, Ar–H), 4.59 (q, J = 21.3 Hz, 2H, OCH2), 1.51 (t, J = 14.1 Hz, 3H, CH3)·13C-NMR (75 MHz, CDCl3) (δ, ppm): 161.08, 160.21, 147.95, 143.43, 134.88, 128.61, 128.32, 126.01, 122.32, 62.16, 13.92. IR νmax(neat): 1728 (C O, ester), 1676 (C O, amide) cm−1. LC-MS: (m/z) [M + H]+ 219.03.
3.2.2. Synthesis of ethyl 4-oxo-3-(prop-2-ynyl)-3,4-dihydroquinazoline-2-carboxylate (2)
To a solution of ethyl 4-oxo-3,4-dihydroquinazoline-2-carboxylate (1, 5 g, 1 eq.) in anhydrous DMF (25 mL) at 0 °C was added K2CO3 (6.33 g, 2 eq.) and the reaction mixture was stirred for 15 min. Propargyl bromide (2.45 mL, 1.2 eq., 80 wt% in toluene) was added dropwise to the mixture and stirred at 0 °C for 1 h. The reaction mixture was then allowed to attain room temperature. After removal of DMF under vacuum, the residue was dissolved in ethyl acetate. The organic layer was washed with a saturated citric acid solution, water and brine, dried over anhydrous Na2SO4 and concentrated under reduced pressure to afford an oily residue. Column purification gave the product (2) as colorless crystals in excellent yield.
Colorless crystalline solid, mp: 92–94 °C, yield: 92%, Rf (30% EtOAc in cyclohexane): 0.6, anal (C14H12N2O3) calc. C 65.62 H 4.72 N 10.93, found: C 65.58 H 4.67 N 10.88. 1H-NMR (300 MHz, CDCl3) (δ, ppm): 8.33 (d, J = 8.1 Hz, 1H, Ar–H), 7.82–7.77 (m, 2H, Ar–H), 7.63–7.56 (m, 1H, Ar–H), 5.16 (s, 2H, CH2), 4.55 (q, J = 21.3 Hz, 2H, OCH2), 2.32 (s, 1H, CH), 1.49 (t, J = 14.4 Hz, 3H, CH3). 13C-NMR (75 MHz, CDCl3) (δ, ppm): 161.20, 160.34, 145.94, 145.45, 134.88, 128.79, 128.31, 127.23, 121.75, 77.72, 73.21, 63.42, 33.11, 13.98. IR νmax(neat): 3207 ( CH), 2127 (C C), 1732 (C O, ester) and 1689 (C O, amide) cm−1. LC-MS: (m/z) [M + H]+ 257.2.
3.2.3. General procedure for the synthesis of azides (4a–4q)
To a stirred solution of aniline/substituted anilines, 3a–q (0.2 g, 1 eq.) in ethyl acetate (3–4 mL) concentrated HCl (0.6 mL) was added at 0 °C, then a solution of NaNO2 (1.2 eq.) in H2O (2.7 mL) was added. After stirring for a further 1 h at 0–5 °C, a solution of NaN3 (1.2 eq.) in H2O (2.7 mL) was added to the above reaction mixture, stirred and allowed to proceed at room temperature till completion of the reaction monitored by TLC. H2O was added and the azide derivatives were extracted with ethyl acetate, treated with brine, dried over anhydrous Na2SO4 and concentrated under reduced pressure. The crude product was directly used for the next step without further purification.
3.2.4. General procedure for the synthesis of compounds (5a–q)
To a solution of tetrahydrofuran (3 mL) and water (6 mL), ethyl 4-oxo-3-(prop-2-ynyl)-3,4-dihydroquinazoline-2-carboxylate (2, 1 eq.) was added while stirring. After thorough mixing, phenyl azide/substituted phenyl azides (4a–q, 1 eq.), CuSO4·5H2O (0.174 eq.) and sodium ascorbate (0.522 eq.) were added separately. The reaction mixture was stirred at room temperature till completion of the reaction, then diluted with H2O and extracted with ethyl acetate. The combined organic phases were treated with brine, dried over anhydrous Na2SO4, filtered and concentrated under reduced pressure to afford a crude product which was purified by column chromatography using ethyl acetate and hexane as the solvent system. The physico-chemical properties of compounds (5a–q) have been listed in Table S13 (ESI†).
Ethyl 4-oxo-3-((1-phenyl-1H-1,2,3-triazol-4-yl)methyl)-3,4-dihydroquinazoline-2-carboxylate (5a)
Off-white solid, Rf (30% EtOAc in cyclohexane): 0.31, anal (C20H17N5O3) calc. C 63.99 H 4.56 N 18.66, found: C 63.97 H 4.51 N 18.62. 1H-NMR (300 MHz, CDCl3) (δ, ppm): 8.32 (d, J = 7.8 Hz, 1H, Ar–H), 8.1 (s, 1H, CH), 7.79 (d, J = 3.6 Hz, 1H, Ar–H), 7.74–7.67 (m, 2H, Ar–H), 7.61–7.39 (m, 5H, Ar–H), 5.67 (s, 2H, CH2), 4.59 (q, J = 12.6 Hz, 2H, OCH2), 1.47 (t, J = 14.1 Hz, 3H, CH3). 13C-NMR (75 MHz, CDCl3) (δ, ppm): 161.49, 161.16, 146.42, 146.27, 136.86, 134.85, 129.72, 128.87, 128.52, 128.19, 126.90, 121.82, 120.56, 63.74, 39.56, 13.95. IR νmax(neat): 3080 ( CH), 1728 (C O, ester) and 1687 (C O, amide) cm−1. LC-MS: (m/z) [M + H]+ 376.3.
Ethyl 3-((1-(2-chlorophenyl)-1H-1,2,3-triazol-4-yl)methyl)-4-oxo-3,4-dihydroquinazoline-2-carboxylate (5b)
Off-white solid, Rf (5% methanol in DCM): 0.68, anal (C20H16ClN5O3) calc. C 58.61 H 3.94 Cl 8.65 N 17.09, found: C 58.56 H 3.92 Cl 8.61 N 17.06. 1H-NMR (300 MHz, CDCl3) (δ, ppm): 8.31 (d, J = 8.1 Hz, 1H, Ar–H), 8.09 (s, 1H, CH), 7.79 (d, J = 4.2 Hz, 2H, Ar–H), 7.57–7.53 (m, 3H, Ar–H), 7.44–7.39 (m, 2H, Ar–H), 5.71 (s, 2H, CH2), 4.59 (q, J = 14.1 Hz, 2H, OCH2), 1.48 (t, J = 14.4 Hz, 3H, CH3). 13C-NMR (75 MHz, CDCl3) (δ, ppm): 161.53, 161.09, 146.42, 146.26, 142.45, 134.82, 134.72, 130.84, 130.75, 128.65, 128.51, 128.21, 127.86, 127.75, 126.95, 125.51, 121.85, 63.72, 39.34, 13.96. IR νmax(neat): 3090 ( CH), 1743 (C O, ester) and 1663 (C O, amide) cm−1. LC-MS: (m/z) [M + H]+ 410.1.
Ethyl 3-((1-(2-fluorophenyl)-1H-1,2,3-triazol-4-yl)methyl)-4-oxo-3,4-dihydroquinazoline-2-carboxylate (5c)
Colorless crystalline solid, Rf (30% EtOAc in cyclohexane): 0.34, anal (C20H16FN5O3) calc. C 61.07 H 4.10 F 4.83 N 17.80, found: C 61.04 H 4.06 F 4.79 N 17.78. 1H-NMR (300 MHz, CDCl3) (δ, ppm): 8.32 (d, J = 7.8 Hz, 1H, Ar–H), 8.18 (s, 1H, CH), 7.90–7.79 (m, 2H, Ar–H), 7.59–7.55 (m, 1H, Ar–H), 7.43–7.28 (m, 4H, Ar–H), 5.70 (s, 2H, CH2), 4.59 (q, J = 21.3 Hz, 2H, OCH2), 1.48 (t, J = 6.9 Hz, 3H, CH3). 13C-NMR (75 MHz, CDCl3) (δ, ppm): 161.54, 161.09, 146.37, 146.26, 143.06, 134.81, 130.39, 130.29, 128.51, 128.20, 128.11, 127.66, 126.97, 125.65, 124.64, 121.86, 116.87, 63.70, 39.31, 13.93. IR νmax(neat): 3030 ( CH), 1739 (C O, ester) and 1674 (C O, amide) cm−1. LC-MS: (m/z) [M + H]+ 394.3.
Ethyl 4-oxo-3-((1-o-tolyl-1H-1,2,3-triazol-4-yl)methyl)-3,4-dihydroquinazoline-2-carboxylate (5d)
White solid, Rf (5% methanol in DCM): 0.81, anal (C21H19N5O3) calc. C 64.77 H 4.92 N 17.98, found: C 64.75 H 4.88 N 17.95. 1H-NMR (300 MHz, CDCl3) (δ, ppm): 8.32 (d, J = 7.5 Hz, 1H, Ar–H), 8.06–7.79 (m, 3H, Ar–H), 7.57 (s, 1H, CH), 7.34–7.14 (m, 4H, Ar–H), 5.69 (s, 2H, CH2), 4.59 (q, J = 21.6 Hz, 2H, OCH2), 2.45 (s, 3H, CH3), 1.49 (t, J = 6.3 Hz, 3H, CH3). 13C-NMR (75 MHz, CDCl3) (δ, ppm): 161.53, 161.12, 146.32, 142.52, 136.29, 134.81, 134.61, 131.46, 129.89, 128.48, 128.22, 128.11, 127.66, 126.95, 125.91, 124.91, 121.88, 63.71, 39.43, 22.63, 13.96. IR νmax(neat): 3065 ( CH), 1736 (C O, ester) and 1676 (C O, amide) cm−1. LC-MS: (m/z) [M + H]+ 390.2.
Ethyl 3-((1-(2-methoxyphenyl)-1H-1,2,3-triazol-4-yl)methyl)-4-oxo-3,4-dihydroquinazoline-2-carboxylate (5e)
White solid, Rf (5% methanol in DCM): 0.60, anal (C21H19N5O4) calc. C 62.22 H 4.72 N 17.27, found: C 62.19 H 4.67 N 17.24. 1H-NMR (300 MHz, CDCl3) (δ, ppm): 8.31 (d, J = 7.8 Hz, 1H, Ar–H), 8.19 (s, 1H, CH), 7.79 (d, J = 3.6 Hz, 2H, Ar–H), 7.70 (d, J = 1.5 Hz, 1H, Ar–H), 7.58–7.53 (m, 1H, Ar–H), 7.43–7.37 (m, 1H, Ar–H), 7.08–7.04 (m, 2H, Ar–H), 5.72 (s, 2H, CH2), 4.59 (q, J = 21.3 Hz, 2H, OCH2), 3.86 (s, 3H, CH3), 1.47 (t, J = 14.1 Hz, 3H, CH3). 13C-NMR (75 MHz, CDCl3) (δ, ppm): 161.59, 161.09, 151.10, 146.59, 146.27, 141.99, 134.74, 130.19, 128.42, 128.17, 126.94, 126.09, 125.48, 121.89, 121.13, 121.15, 63.67, 55.93, 39.22, 13.93. IR νmax(neat): 3075 ( CH), 1739 (C O, ester) and 1678 (C O, amide) cm−1. LC-MS: (m/z) [M + H]+ 406.2.
Ethyl 3-((1-(2-nitrophenyl)-1H-1,2,3-triazol-4-yl)methyl)-4-oxo-3,4-dihydroquinazoline-2-carboxylate (5f)
Yellow solid, Rf (5% methanol in DCM): 0.69, anal (C20H16N6O5) calc. C 57.14 H 3.84 N 19.99, found: C 57.11 H 3.82 N 19.93. 1H-NMR (300 MHz, CDCl3) (δ, ppm): 8.29 (d, J = 8.1 Hz, 1H, Ar–H), 8.08–8.05 (m, 1H, Ar–H), 8.00 (s, 1H, CH), 7.81–7.66 (m, 4H, Ar–H), 7.61–7.54 (m, 2H, Ar–H), 5.68 (s, 2H, CH2), 4.59 (q, J = 9.9 Hz, 2H, OCH2), 1.49 (t, J = 7.2 Hz, 3H, CH3). 13C-NMR (75 MHz, CDCl3) (δ, ppm): 161.48, 161.14, 146.36, 146.28, 144.39, 143.20, 134.88, 133.80, 130.88, 130.08, 128.55, 128.25, 128.12, 127.90, 126.91, 125.63, 125.13, 121.82, 63.83, 39.42, 13.93. IR νmax(neat): 3045 ( CH), 1739 (C O, ester) and 1670 (C O, amide) cm−1. LC-MS: (m/z) [M + H]+ 421.3.
Ethyl 3-((1-(3-chlorophenyl)-1H-1,2,3-triazol-4-yl)methyl)-4-oxo-3,4-dihydroquinazoline-2-carboxylate (5g)
White solid, Rf (5% m ethanol in DCM): 0.86, anal (C20H16ClN5O3) calc. C 58.61 H 3.94 Cl 8.65 N 17.09, found: C 58.58 H 3.90 Cl 8.61 N 17.06. 1H-NMR (300 MHz, CDCl3) (δ, ppm): 8.31 (d, J = 7.8 Hz, 1H, Ar–H), 8.12 (s, 1H, CH), 7.83–7.74 (m, 3H, Ar–H), 7.61–7.54 (m, 1H, Ar–H), 7.46–7.37 (m, 2H, Ar–H), 7.19–7.12 (m, 1H, Ar–H), 5.65 (s, 2H, CH2), 4.59 (q, J = 21.6 Hz, 2H, OCH2), 1.49 (t, J = 7.2 Hz, 3H, CH3). 13C-NMR (75 MHz, CDCl3) (δ, ppm): 161.47, 161.18, 146.26, 143.65, 137.67, 135.58, 134.91, 130.81, 128.93, 128.58, 128.23, 128.12, 127.66, 126.90, 125.66, 121.79, 63.78, 39.58, 13.97. IR νmax(neat): 3091 ( CH), 1739 (C O, ester) and 1663 (C O, amide) cm−1. LC-MS: (m/z) [M + H]+ 410.1.
Ethyl 3-((1-(3-fluorophenyl)-1H-1,2,3-triazol-4-yl)methyl)-4-oxo-3,4-dihydroquinazoline-2-carboxylate (5h)
White solid, Rf (5% methanol in DCM): 0.52, anal (C20H16FN5O3) calc. C 61.07 H 4.10 F 4.83 N 17.80, found: C 61.05 H 4.08 F 4.77 N 17.76. 1H-NMR (300 MHz, CDCl3) (δ, ppm): 8.31 (d, J = 7.8 Hz, 1H, Ar–H), 8.11 (s, 1H, CH), 7.79 (d, J = 3.9 Hz, 1H, Ar–H), 7.59–7.42 (m, 4H, Ar–H), 7.19–7.10 (m, 2H, Ar–H), 5.66 (s, 2H, CH2), 4.59 (q, J = 21.3 Hz, 2H, OCH2), 1.48 (t, J = 14.1 Hz, 3H, CH3). 13C-NMR (75 MHz, CDCl3) (δ, ppm): 161.48, 161.39, 146.33, 146.28, 143.62, 137.92, 134.89, 130.23, 131.11, 128.57, 128.24, 128.11, 127.66, 126.90, 121.81, 121.70, 115.93, 63.76, 39.57, 13.96.IR νmax(neat): 3069 ( CH), 1737 (C O, ester) and 1657 (C O, amide) cm−1. LC-MS: (m/z) [M + H]+ 394.2.
Ethyl 4-oxo-3-((1-m-tolyl-1H-1,2,3-triazol-4-yl)methyl)-3,4-dihydroquinazoline-2-carboxylate (5i)
White solid, Rf (5% methanol in DCM): 0.67, anal (C21H19N5O3) calc. C 64.77 H 4.92 N 17.98, found: C 64.74 H 4.87 N 17.96. 1H-NMR (300 MHz, CDCl3) (δ, ppm): 8.32 (d, J = 8.1 Hz, 1H, Ar–H), 8.07 (s, 1H, CH), 7.79 (d, J = 3.6 Hz, 2H, Ar–H), 7.59–7.44 (m, 3H, Ar–H), 7.38–7.11 (m, 2H, Ar–H), 5.66 (s, 2H, CH2), 4.59 (q, J = 21.3 Hz, 2H, OCH2), 2.41 (s, 3H, CH3), 1.47 (t, J = 14.1 Hz, 3H, CH3). 13C-NMR (75 MHz, CDCl3) (δ, ppm): 161.50, 161.16, 146.47, 146.30, 143.25, 139.96, 136.80, 134.83, 129.60, 128.20, 128.11, 127.66, 126.91, 125.65, 121.84, 121.72, 121.19, 63.73, 39.54, 22.64, 13.96. IR νmax(neat): 3060 ( CH), 1737 (C O, ester) and 1657 (C O, amide) cm−1. LC-MS: (m/z) [M + H]+ 390.2.
Ethyl 3-((1-(3-methoxyphenyl)-1H-1,2,3-triazol-4-yl)methyl)-4-oxo-3,4-dihydroquinazoline-2-carboxylate (5j)
White solid, Rf (5% methanol in DCM): 0.44, anal (C21H19N5O4) calc. C 62.22 H 4.72 N 17.27, found: C 62.17 H 4.66 N 17.25. 1H-NMR (300 MHz, CDCl3) (δ, ppm): 8.31 (d, J = 8.1 Hz, 1H, Ar–H), 8.12 (s, 1H, CH), 7.80–7.75 (m, 2H, Ar–H), 7.61–7.54 (m, 2H, Ar–H), 7.46–7.37 (m, 2H, Ar–H), 7.19–7.12 (m, 1H, Ar–H), 5.66 (s, 2H, CH2), 4.60 (q, J = 21.3 Hz, 2H, OCH2), 3.86 (s, 3H, CH3), 1.48 (t, J = 14.4 Hz, 3H, CH3). 13C-NMR (75 MHz, CDCl3) (δ, ppm): 161.49, 161.16, 160.56, 146.42, 146.28, 143.29, 137.85, 134.86, 130.49, 128.53, 128.21, 127.66, 126.91, 121.80, 114.83, 112.45, 106.28, 63.74, 55.62, 39.52, 13.97. IR νmax(neat): 3090 ( CH), 1743 (C O, ester) and 1663 (C O, amide) cm−1. LC-MS: (m/z) [M + H]+ 406.1.
Ethyl 3-((1-(3-nitrophenyl)-1H-1,2,3-triazol-4-yl)methyl)-4-oxo-3,4-dihydroquinazoline-2-carboxylate (5k)
White solid, Rf (5% methanol in DCM): 0.72, anal (C20H16N6O5) calc. C 57.14 H 3.84 N 19.99, found: C 57.10 H 3.82 N 19.95. 1H-NMR (300 MHz, CDCl3) (δ, ppm): 8.58 (d, J = 1.5 Hz, 1H, Ar–H), 8.34–8.27 (m, 2H, Ar–H), 8.25 (s, 1H, CH), 8.15–8.11 (m, 1H, Ar–H), 7.82–7.56 (m, 4H, Ar–H), 5.67 (s, 2H, CH2), 4.61 (q, J = 21.3 Hz, 2H, OCH2), 1.49 (t, J = 14.1 Hz, 3H, CH3). 13C-NMR (75 MHz, CDCl3) (δ, ppm): 161.44, 161.19, 148.91, 146.24, 144.15, 137.56, 134.96, 130.95, 128.66, 128.25, 126.89, 125.89, 123.28, 121.77, 63.78, 39.67, 13.98. IR νmax(neat): 3105 ( CH), 1736 (C O, ester) and 1678 (C O, amide) cm−1. LC-MS: (m/z) [M + H]+ 421.2.
Ethyl 3-((1-(4-chlorophenyl)-1H-1,2,3-triazol-4-yl)methyl)-4-oxo-3,4-dihydroquinazoline-2-carboxylate (5l)
Light yellow solid, Rf (5% methanol in DCM): 0.76, anal (C20H16ClN5O3) calc. C 58.61 H 3.94 Cl 8.65 N 17.09, found: C 58.56 H 3.91 Cl 8.63 N 17.05. 1H-NMR (300 MHz, CDCl3) (δ, ppm): 8.31 (d, J = 7.8 Hz, 1H, Ar–H), 8.11 (s, 1H, CH), 7.79 (d, J = 3.6 Hz, 2H, Ar–H), 7.65 (d, J = 8.7 Hz, 2H, Ar–H), 7.60–7.54 (m, 1H, Ar–H), 7.48–7.45 (m, 2H, Ar–H), 5.65 (s, 2H, CH2), 4.60 (q, J = 20.7 Hz, 2H, OCH2), 1.48 (t, J = 14.1 Hz, 3H, CH3). 13C-NMR (75 MHz, CDCl3) (δ, ppm): 161.47, 161.18, 146.26, 135.34, 134.91, 134.67, 129.92, 128.58, 128.23, 126.89, 121.70, 63.77, 39.59, 13.97. IR νmax(neat): 3052 ( CH), 1739 (C O, ester) and 1657 (C O, amide) cm−1. LC-MS: (m/z) [M + H]+ 410.1.
Ethyl 3-((1-(4-fluorophenyl)-1H-1,2,3-triazol-4-yl)methyl)-4-oxo-3,4-dihydroquinazoline-2-carboxylate (5m)
White crystalline solid, Rf (5% methanol in DCM): 0.69, anal (C20H16FN5O3) calc. C 61.07 H 4.10 F 4.83 N 17.80, found: C 61.06 H 4.07 F 4.80 N 17.75. 1H-NMR (300 MHz, CDCl3) (δ, ppm): 8.31 (d, J = 7.8 Hz, 1H, Ar–H), 8.06 (s, 1H, CH), 7.79 (d, J = 3.9 Hz, 2H, Ar–H), 7.68–7.64 (m, 2H, Ar–H), 7.61–7.53 (m, 1H, Ar–H), 7.29–7.12 (m, 2H, Ar–H), 5.66 (s, 2H, CH2), 4.59 (q, J = 21.6 Hz, 2H, OCH2), 1.48 (t, J = 14.1 Hz, 3H, CH3). 13C-NMR (75 MHz, CDCl3) (δ, ppm): 161.48, 161.18, 146.38, 146.29, 143.50, 134.88, 133.17, 128.55, 128.23, 128.11, 127.66, 121.82, 63.75, 39.57, 13.96. IR νmax(neat): 3067 ( CH), 1739 (C O, ester) and 1657 (C O, amide) cm−1. LC-MS: (m/z) [M + H]+ 394.3.
Ethyl 4-oxo-3-((1-p-tolyl-1H-1,2,3-triazol-4-yl)methyl)-3,4-dihydroquinazoline-2-carboxylate (5n)
Off-white solid, Rf (5% methanol in DCM): 0.86, anal (C21H19N5O3) calc. C 64.77 H 4.92 N 17.98, found: C 64.76 H 4.89 N 17.96. 1H-NMR (300 MHz, CDCl3) (δ, ppm): 8.31 (d, J = 7.8 Hz, 1H, Ar–H), 8.06 (s, 1H, CH), 7.79 (d, J = 3.6 Hz, 2H, Ar–H), 7.59–7.54 (m, 3H, Ar–H), 7.28 (d, J = 3.6 Hz, 2H, Ar–H), 5.66 (s, 2H, CH2), 4.59 (q, J = 21.6 Hz, 2H, OCH2), 2.39 (s, 3H, CH3), 1.47 (t, J = 14.4 Hz, 3H, CH3). 13C-NMR (75 MHz, CDCl3) (δ, ppm): 161.49, 161.14, 146.47, 146.29, 138.98, 134.81, 134.59, 130.19, 128.48, 128.19, 126.91, 121.84, 121.66, 120.46, 63.71, 39.53, 21.06, 13.94. IR νmax(neat): 3047 ( CH), 1741 (C O, ester) and 1663 (C O, amide) cm−1. LC-MS: (m/z) [M + H]+ 390.2.
Ethyl 3-((1-(4-methoxyphenyl)-1H-1,2,3-triazol-4-yl)methyl)-4-oxo-3,4-dihydroquinazoline-2-carboxylate (5o)
White solid, Rf (5% methanol in DCM): 0.76. Anal (C21H19N5O4) calc. C 62.22 H 4.72 N 17.27, found: C 62.17 H 4.68 N 17.25. 1H-NMR (300 MHz, CDCl3) (δ, ppm): 8.31 (d, J = 7.8 Hz, 1H, Ar–H), 8.01 (s, 1H, CH), 7.79 (d, J = 3.6 Hz, 2H, Ar–H), 7.59–7.54 (m, 3H, Ar–H), 7.19–7.12 (m, 1H, Ar–H), 6.99–6.96 (m, 1H, Ar–H), 5.66 (s, 2H, CH2), 4.59 (q, J = 21.3 Hz, 2H, OCH2), 3.84 (s, 3H, CH3), 1.47 (t, J = 14.4 Hz, 3H, CH3). 13C-NMR (75 MHz, CDCl3) (δ, ppm): 161.51, 161.16, 159.88, 146.48, 146.30, 143.14, 134.82, 130.32, 128.49, 128.20, 128.11, 127.66, 126.91, 125.64, 122.20, 121.81, 114.73, 63.72, 55.60, 39.53, 22.65, 13.95. IR νmax(neat): 3090 ( CH), 1739 (C O, ester) and 1659 (C O, amide) cm−1. LC-MS: (m/z) [M + H]+ 406.2.
Ethyl 3-((1-(4-nitrophenyl)-1H-1,2,3-triazol-4-yl)methyl)-4-oxo-3,4-dihydroquinazoline-2-carboxylate (5p)
Yellow solid, Rf (5% methanol in DCM): 0.70, anal (C20H16N6O5) calc. C 57.14 H 3.84 N 19.99, found: C 57.11 H 3.82 N 19.96. 1H-NMR (300 MHz, CDCl3) (δ, ppm): 8.38 (dd, J = 9.3 Hz, 1H, Ar–H), 8.31 (s, 1H, CH), 8.28 (d, J = 5.4 Hz, 2H, Ar–H), 7.95 (dd, J = 9.0 Hz, 2H, Ar–H), 7.84–7.74 (m, 2H, Ar–H), 7.60–7.55 (m, 1H, Ar–H), 5.66 (s, 2H, CH2), 4.60 (q, J = 21.3 Hz, 2H, OCH2), 1.49 (t, J = 14.4 Hz, 3H, CH3). 13C-NMR (75 MHz, CDCl3) (δ, ppm): 161.44, 161.21, 147.30, 146.23, 143.15, 140.97, 134.99, 128.68, 128.27, 126.85, 125.50, 121.75, 120.53, 63.80, 39.67, 13.98. IR νmax(neat): 3093 ( CH), 1734 (C O, ester) and 1654 (C O, amide) cm−1. LC-MS: (m/z) [M + H]+ 421.3.
4-(4-((2-(ethoxycarbonyl)-4-oxoquinazolin-3(4H)-yl)methyl)-1H-1,2,3-triazol-1-yl)benzoic acid (5q)
White solid, Rf (5% methanol in DCM): 0.14, anal (C21H17N5O5) calc. C 60.14 H 4.09 N 16.70, found: C 60.10 H 4.07 N 16.66. 1H-NMR (300 MHz, DMSO-d6) (δ, ppm): 8.87 (s, 1H, CH), 8.21 (d, J = 8.1 Hz, 1H, Ar–H), 8.10–7.98 (m, 4H, Ar–H), 7.93–7.88 (m, 1H, Ar–H), 7.79–7.76 (m, 1H, Ar–H), 7.68–7.63 (m, 1H, Ar–H), 5.49 (s, 2H, CH2), 4.35 (q, J = 21.3 Hz, 2H, OCH2), 1.19 (t, J = 14.1 Hz, 3H, CH3). 13C-NMR (75 MHz, DMSO-d6) (δ, ppm): 166.82, 161.29, 160.64, 146.92, 146.23, 144.50, 139.79, 135.61, 131.52, 131.17, 129.18, 128.18, 127.07, 122.30, 121.98, 120.12, 63.39, 39.13, 14.02. IR νmax(neat): 3067 ( CH), 1739 (C O, ester), 1695 (C O, acid) and 1657 (C O, amide) cm−1. LC-MS: (m/z) [M + H]+ 420.1.
3.2.4. General procedure for synthesis of compounds (6a–q)
To the stirred solution of esters 5a–q (1.0 eq.) separately in the mixture of THF (3 mL) and water (1 mL) was added lithium hydroxide monohydrate (3.0 eq.) and the reaction mixture was stirred at room temperature. After completion of reaction (monitored by TLC) the reaction mixture was concentrated under reduced pressure, water was added, acidified with aqueous 1 N HCl solution to make acidic pH and extracted with ethyl acetate. The combined organic layer was washed with brine solution, dried over anhydrous Na2SO4, filtered and concentrated under reduced pressure to afford the corresponding acids in good yield. The physico-chemical properties of compounds (6a–q) have been listed in Table S14 (ESI†).
4-Oxo-3-((1-phenyl-1H-1,2,3-triazol-4-yl)methyl)-3,4-dihydroquinazoline-2-carboxylic acid (6a)
Off-white solid, Rf (5% methanol in DCM): 0.55, anal (C18H13N5O3) calc. C 62.24 H 3.77 N 20.16, found: C 62.20 H 3.75 N 20.14. 1H-NMR (300 MHz, CDCl3) (δ, ppm): 8.44 (s, 1H, Ar–H), 8.29 (d, J = 8.4 Hz, 1H, Ar–H), 8.22 (s, 1H, CH), 7.79–7.69 (m, 3H, Ar–H), 7.53–7.40 (m, 4H, Ar–H), 5.37 (s, 2H, CH2). 13C-NMR (75 MHz, CDCl3) (δ, ppm): 161.06, 148.13, 146.31, 142.91, 136.79, 134.49, 129.76, 129.00, 127.65, 127.44, 126.54, 122.10, 121.97, 120.62, 41.78. IR νmax(neat): 3064 ( CH), 1721 (C O, acid) and 1674 (C O, amide) cm−1. LC-MS: (m/z) [M − H]− 346.0.
3-((1-(2-Chlorophenyl)-1H-1,2,3-triazol-4-yl)methyl)-4-oxo-3,4-dihydroquinazoline-2-carboxylic acid (6b)
White solid, Rf (5% methanol in DCM): 0.44, anal (C18H12ClN5O3) calc. C 56.63 H 3.17 Cl 9.29 N 18.34, found: C 56.61 H 3.14 Cl 9.25 N 18.30. 1H-NMR (300 MHz, CDCl3) (δ, ppm): 8.43 (s, 1H, Ar–H), 8.29 (d, J = 8.4 Hz, 2H, Ar–H), 8.19 (s, 1H, CH), 7.79–7.72 (m, 2H, Ar–H), 7.58–7.39 (m, 3H, Ar–H), 5.39 (s, 2H, CH2). 13C-NMR (75 MHz, CDCl3) (δ, ppm): 164.16, 160.99, 159.95, 148.15, 146.28, 141.97, 134.64, 134.46, 130.93, 130.78, 128.64, 127.90, 127.75, 127.67, 127.42, 126.57, 125.91, 122.01, 41.66. IR νmax(neat): 3071 ( CH), 1721 (C O, acid) and 1678 (C O, amide) cm−1. LC-MS: (m/z) [M − H]− 379.8.
3-((1-(2-Fluorophenyl)-1H-1,2,3-triazol-4-yl)methyl)-4-oxo-3,4-dihydroquinazoline-2-carboxylic acid (6c)
White solid, Rf (5% methanol in DCM): 0.23, anal (C18H12FN5O3) calc. C 59.18 H 3.31 F 5.20 N 19.17, found: C 59.14 H 3.28 F 5.17 N 19.14. 1H-NMR (300 MHz, CDCl3) (δ, ppm): 8.43 (s, 1H, Ar–H), 8.33–8.30 (m, 1H, Ar–H), 8.29 (s, 1H, CH), 7.93–7.80 (m, 1H, Ar–H), 7.79–7.73 (m, 2H, Ar–H), 7.55–7.27 (m, 3H, Ar–H), 5.39 (s, 2H, CH2). 13C-NMR (75 MHz, CDCl3) (δ, ppm): 164.20, 161.05, 148.12, 146.27, 143.04, 134.53, 133.07, 127.68, 127.47, 126.52, 122.67, 122.56, 122.38, 121.95, 116.90, 41.77. IR νmax(neat): 3064 ( CH), 1721 (C O, acid) and 1663 (C O, amide) cm−1. LC-MS: (m/z) [M–COOH]+ 338.0.
4-Oxo-3-((1-o-tolyl-1H-1,2,3-triazol-4-yl)methyl)-3,4-dihydroquinazoline-2-carboxylic acid (6d)
White solid, Rf (5% methanol in DCM): 0.54, anal (C19H15N5O3) calc. C 63.15 H 4.18 N 19.38, found: C 63.11 H 4.15 N 19.37. 1H-NMR (300 MHz, CDCl3) (δ, ppm): 8.43 (s, 1H, Ar–H), 8.29 (d, J = 7.8 Hz, 1H, Ar–H), 7.96 (s, 1H, CH), 7.79–7.72 (m, 2H, Ar–H), 7.54–7.49 (m, 1H, Ar–H), 7.41–7.29 (m, 3H, Ar–H), 5.38 (s, 2H, CH2), 2.12 (s, 3H, CH3). 13C-NMR (75 MHz, CDCl3) (δ, ppm): 161.01, 148.19, 146.31, 142.05, 136.18, 134.44, 133.53, 131.51, 129.97, 127.69, 127.39, 126.84, 126.55, 125.88, 125.30, 122.02, 41.70, 17.89. IR νmax(neat): 3099 ( CH), 1700 (C O, acid) and 1680 (C O, amide) cm−1. LC-MS: (m/z) [M–COOH]+ 334.0.
3-((1-(2-Methoxyphenyl)-1H-1,2,3-triazol-4-yl)methyl)-4-oxo-3,4-dihydroquinazoline-2-carboxylic acid (6e)
White solid, Rf (5% methanol in DCM): 0.41, anal (C19H15N5O4) calc. C 60.47 H 4.01 N 18.56, found: C 60.44 H 3.98 N 18.53. 1H-NMR (300 MHz, CDCl3) (δ, ppm): 8.36 (s, 1H, Ar–H), 8.22 (s, 1H, Ar–H), 8.20 (s, 1H, CH), 7.70–7.62 (m, 2H, Ar–H), 7.44–7.30 (m, 2H, Ar–H), 7.02–6.97 (m, 2H, Ar–H), 5.30 (s, 2H, CH2), 3.79 (s, 3H, CH3). 13C-NMR (75 MHz, CDCl3) (δ, ppm): 160.97, 151.12, 148.19, 146.38, 141.57, 134.35, 130.30, 127.66, 127.30, 126.55, 126.04, 125.87, 125.49, 122.07, 121.16, 112.18, 55.96, 41.54. IR νmax(neat): 3086 ( CH), 1719 (C O, acid) and 1674 (C O, amide) cm−1. LC-MS: (m/z) [M + H]+ 378.2.
3-((1-(2-Nitrophenyl)-1H-1,2,3-triazol-4-yl)methyl)-4-oxo-3,4-dihydroquinazoline-2-carboxylic acid (6f)
Light-yellow solid, Rf (5% methanol in DCM): 0.34, anal (C18H12N6O5) calc. C 55.11 H 3.08 N 21.42, found: C 55.09 H 3.05 N 21.35. 1H-NMR (300 MHz, DMSO-d6) (δ, ppm): 8.41 (s, 1H, Ar–H), 8.35–8.27 (m, 1H, Ar–H), 8.19 (s, 1H, CH), 7.80–7.34 (m, 6H, Ar–H), 5.37 (s, 2H, CH2). 13C-NMR (75 MHz, DMSO-d6) (δ, ppm): 160.41, 148.16, 148.12, 148.30, 144.16, 137.55, 134.91, 131.93, 127.74, 127.63, 126.55, 123.58, 122.81, 122.13, 115.23, 41.48. IR νmax(neat): 3084 ( CH), 1700 (C O, acid) and 1667 (C O, amide) cm−1. LC-MS: (m/z) [M − H]− 390.9.
3-((1-(3-Chlorophenyl)-1H-1,2,3-triazol-4-yl)methyl)-4-oxo-3,4-dihydroquinazoline-2-carboxylic acid (6g)
White solid, Rf (5% methanol in DCM): 0.68, anal (C18H12ClN5O3) calc. C 56.63 H 3.17 Cl 9.29 N 18.34, found: C 56.60 H 3.15 Cl 9.26 N 18.31. 1H-NMR (300 MHz, CDCl3) (δ, ppm): 8.39 (s, 1H, Ar–H), 8.29 (d, J = 8.4 Hz, 1H, Ar–H), 8.22 (s, 1H, CH), 7.80–7.71 (m, 3H, Ar–H), 7.64–7.59 (m, 1H, Ar–H), 7.54–7.39 (m, 2H, Ar–H), 5.35 (s, 2H, CH2). 13C-NMR (75 MHz, CDCl3) (δ, ppm): 161.08, 148.20, 146.12, 143.19, 137.61, 135.63, 134.52, 130.83, 129.06, 127.74, 127.47, 126.52, 122.05, 121.96, 120.86, 118.53, 41.81. IR νmax(neat): 3073 ( CH), 1719 (C O, acid) and 1680 (C O, amide) cm−1. LC-MS: (m/z) [M − H]− 379.8.
3-((1-(3-Fluorophenyl)-1H-1,2,3-triazol-4-yl)methyl)-4-oxo-3,4-dihydroquinazoline-2-carboxylic acid (6h)
White solid, Rf (5% methanol in DCM): 0.66, anal (C18H12FN5O3) calc. C 59.18 H 3.31 F 5.20 N 19.17, found: C 59.16 H 3.29 F 5.18 N 19.15. 1H-NMR (300 MHz, CDCl3) (δ, ppm): 8.39 (s, 1H, Ar–H), 8.29 (d, J = 8.1 Hz, 1H, Ar–H), 8.22 (s, 1H, CH), 7.80–7.72 (m, 2H, Ar–H), 7.54–7.44 (m, 3H, Ar–H), 7.18–7.09 (m, 1H, Ar–H), 5.36 (s, 2H, CH2). 13C-NMR (75 MHz, CDCl3) (δ, ppm): 161.04, 160.58, 148.21, 146.20, 142.87, 136.80, 133.45, 129.51, 126.70, 126.41, 126.33, 122.11, 121.99, 114.94, 112.19, 41.77. IR νmax(neat): 3062 ( CH), 1721 (C O, acid) and 1669 (C O, amide) cm−1. LC-MS: (m/z) [M–COOH]+ 338.1.
4-Oxo-3-((1-m-tolyl-1H-1,2,3-triazol-4-yl)methyl)-3,4-dihydroquinazoline-2-carboxylic acid (6i)
White solid, Rf (5% methanol in DCM): 0.55, anal (C19H15N5O3) calc. C 63.15 H 4.18 N 19.38, found: C 63.12 H 4.14 N 19.35. 1H-NMR (300 MHz, CDCl3) (δ, ppm): 8.41 (s, 1H, Ar–H), 8.29 (d, J = 7.8 Hz, 1H, Ar–H), 8.19 (s, 1H, CH), 7.79–7.68 (m, 2H, Ar–H), 7.53–7.47 (m, 3H, Ar–H), 7.39–7.34 (m, 1H, Ar–H), 5.29 (s, 2H, CH2), 2.36 (s, 3H, CH3). 13C-NMR (75 MHz, CDCl3) (δ, ppm): 161.06, 148.20, 146.24, 142.82, 140.01, 136.74, 134.46, 129.73, 129.52, 127.71, 127.41, 126.53, 122.08, 122.00, 121.25, 117.68, 41.79, 21.35. IR νmax(neat): 3078 ( CH), 1721 (C O, acid) and 1680 (C O, amide) cm−1. LC-MS: (m/z) [M–COOH]+ 334.0.
3-((1-(3-Methoxyphenyl)-1H-1,2,3-triazol-4-yl)methyl)-4-oxo-3,4-dihydroquinazoline-2-carboxylic acid (6j)
White solid, Rf (5% methanol in DCM): 0.59, anal (C19H15N5O4) calc. C 60.47 H 4.01 N 18.56, found: C 60.46 H 3.97 N 18.54. 1H-NMR (300 MHz, CDCl3) (δ, ppm): 8.33 (s, 1H, Ar–H), 8.21 (d, J = 7.8 Hz, 1H, Ar–H), 8.11 (s, 1H, CH), 7.71–7.63 (m, 2H, Ar–H), 7.45–7.40 (m, 1H, Ar–H), 7.33–7.27 (m, 1H, Ar–H), 7.21–7.14 (m, 1H, Ar–H), 6.89–6.86 (m, 1H, Ar–H), 5.27 (s, 2H, CH2), 3.77 (s, 3H, CH3). 13C-NMR (75 MHz, CDCl3) (δ, ppm): 161.04, 160.59, 148.20, 146.22, 142.88, 137.80, 134.45, 130.51, 127.70, 127.41, 126.53, 122.11, 121.99, 114.95, 112.49, 106.34, 55.63, 41.74. IR νmax(neat): 3069 ( CH), 1721 (C O, acid) and 1665 (C O, amide) cm−1. LC-MS: (m/z) [M + H]+ 378.2.
3-((1-(3-Nitrophenyl)-1H-1,2,3-triazol-4-yl)methyl)-4-oxo-3,4-dihydroquinazoline-2-carboxylic acid (6k)
White solid, Rf (5% methanol in DCM): 0.54, anal (C18H12N6O5) calc. C 55.11 H 3.08 N 21.42, found: C 55.08 H 3.06 N 21.38. 1H-NMR (300 MHz, DMSO-d6) (δ, ppm): 8.69 (d, J = 1.8 Hz, 1H, Ar–H), 8.61 (s, 1H, CH), 8.38 (dd, J = 10.2 Hz, 1H, Ar–H), 8.29 (dd, J = 10.5 Hz, 1H, Ar–H), 8.16 (d, J = 7.8 Hz, 2H, Ar–H), 7.89–7.82 (m, 1H, Ar–H), 7.72–7.69 (m, 1H, Ar–H), 7.58–7.53 (m, 1H, Ar–H), 5.40 (s, 2H, CH2). 13C-NMR (75 MHz, DMSO-d6) (δ, ppm): 160.43, 148.96, 148.42, 148.31, 144.68, 137.55, 134.92, 131.94, 127.75, 127.64, 126.55, 123.59, 122.82, 122.13, 115.24, 41.49. IR νmax(neat): 3069 ( CH), 1709 (C O, acid) and 1657 (C O, amide) cm−1. LC-MS: (m/z) [M − H]− 390.9.
3-((1-(4-Chlorophenyl)-1H-1,2,3-triazol-4-yl)methyl)-4-oxo-3,4-dihydroquinazoline-2-carboxylic acid (6l)
Off-white solid, Rf (5% methanol in DCM): 0.58, anal (C18H12ClN5O3) calc. C 56.63 H 3.17 Cl 9.29 N 18.34, found: C 56.60 H 3.15 Cl 9.26 N 18.31. 1H-NMR (300 MHz, CDCl3) (δ, ppm): 8.40 (s, 1H, Ar–H), 8.28 (d, J = 7.8 Hz, 1H, Ar–H), 8.19 (s, 1H, CH), 7.79–7.64 (m, 4H, Ar–H), 7.54–7.46 (m, 2H, Ar–H), 5.29 (s, 2H, CH2). 13C-NMR (75 MHz, CDCl3) (δ, ppm): 161.06, 148.16, 146.17, 143.16, 135.27, 134.83, 134.52, 129.95, 127.71, 127.47, 126.51, 121.99, 121.75, 41.79. IR νmax(neat): 3065 ( CH), 1721 (C O, acid) and 1672 (C O, amide) cm−1. LC-MS: (m/z) [M − H]− 379.8.
3-((1-(4-Fluorophenyl)-1H-1,2,3-triazol-4-yl)methyl)-4-oxo-3,4-dihydroquinazoline-2-carboxylic acid (6m)
Off-white solid, Rf (5% methanol in DCM): 0.62, anal (C18H12FN5O3) calc. C 59.18 H 3.31 F 5.20 N 19.17, found: C 59.15 H 3.29 F 5.18 N 19.13. 1H-NMR (300 MHz, CDCl3) (δ, ppm): 8.42 (s, 1H, Ar–H), 8.29 (d, J = 7.5 Hz, 1H, Ar–H), 8.18 (s, 1H, CH), 7.76–7.66 (m, 2H, Ar–H), 7.54–7.48 (m, 1H, Ar–H), 7.22–7.12 (m, 1H, Ar–H), 5.36 (s, 2H, CH2). 13C-NMR (75 MHz, CDCl3) (δ, ppm): 164.21, 161.05, 148.13, 146.27, 143.05, 134.52, 133.08, 127.69, 127.47, 126.52, 122.68, 122.57, 122.28, 121.96, 116.90, 41.79. IR νmax(neat): 3065 ( CH), 1719 (C O, acid) and 1667 (C O, amide) cm−1. LC-MS: (m/z) [M–COOH]+ 338.1.
4-Oxo-3-((1-p-tolyl-1H-1,2,3-triazol-4-yl)methyl)-3,4-dihydroquinazoline-2-carboxylic acid (6n)
White solid, Rf (5% methanol in DCM): 0.50, anal (C19H15N5O3) calc. C 63.15 H 4.18 N 19.38, found: C 63.12 H 4.15 N 19.34. 1H-NMR (300 MHz, CDCl3) (δ, ppm): 8.41 (s, 1H, Ar–H), 8.29 (d, J = 8.4 Hz, 1H, Ar–H), 8.16 (s, 1H, CH), 7.95–7.71 (m, 3H, Ar–H), 7.59–7.48 (m, 3H, Ar–H), 5.36 (s, 2H, CH2), 2.07 (s, 3H, CH3). 13C-NMR (75 MHz, CDCl3) (δ, ppm): 161.04, 148.17, 146.29, 142.77, 139.13, 134.45, 130.23, 127.68, 127.39, 126.53, 122.00, 120.51, 107.63, 106.36, 41.76, 29.50. IR νmax(neat): 3077 ( CH), 1721 (C O, acid) and 1680 (C O, amide) cm−1. LC-MS: (m/z) [M–COOH]+ 334.0.
3-((1-(4-Methoxyphenyl)-1H-1,2,3-triazol-4-yl)methyl)-4-oxo-3,4-dihydroquinazoline-2-carboxylic acid (6o)
White solid, Rf (5% methanol in DCM): 0.61, Anal (C19H15N5O4) calc. C 60.47 H 4.01 N 18.56, found: C 60.45 H 3.99 N 18.55. 1H-NMR (300 MHz, CDCl3) (δ, ppm): 8.41 (s, 1H, Ar–H), 8.29 (d, J = 8.4 Hz, 1H, Ar–H), 8.11 (s, 1H, CH), 7.79–7.72 (m, 2H, Ar–H), 7.61–7.49 (m, 2H, Ar–H), 7.01–6.98 (m, 2H, Ar–H), 5.35 (s, 2H, CH2), 3.85 (s, 3H, CH3). 13C-NMR (75 MHz, CDCl3) (δ, ppm): 160.00, 148.20, 146.27, 142.72, 134.46, 130.25, 127.71, 127.41, 126.54, 122.26, 114.78, 107.63, 106.37, 55.61, 41.77. IR νmax(neat): 3075 ( CH), 1717 (C O, acid) and 1674 (C O, amide) cm−1. LC-MS: (m/z) [M + H]+ 378.2.
3-((1-(4-Nitrophenyl)-1H-1,2,3-triazol-4-yl)methyl)-4-oxo-3,4-dihydroquinazoline-2-carboxylic acid (6p)
Off-white solid, Rf (5% methanol in DCM): 0.53, anal (C18H12N6O5) calc. C 55.11 H 3.08 N 21.42, found: C 55.09 H 3.05 N 21.39. 1H-NMR (300 MHz, DMSO-d6) (δ, ppm): 8.61 (s, 1H, CH), 8.41 (d, J = 9.0 Hz, 2H, Ar–H), 8.21–8.14 (m, 3H, Ar–H), 7.87–7.82 (m, 1H, Ar–H), 7.72–7.69 (m, 1H, Ar–H), 7.58–7.53 (m, 1H, Ar–H), 5.40 (s, 2H, CH2). 13C-NMR (75 MHz, DMSO-d6) (δ, ppm): 160.43, 148.41, 148.30, 147.18, 144.86, 141.18, 134.92, 127.74, 127.63, 126.55, 125.94, 122.76, 122.12, 121.06, 41.48. IR νmax(neat): 3069 ( CH), 1719 (C O, acid) and 1665 (C O, amide) cm−1. LC-MS: (m/z) [M − H]− 390.9.
3-((1-(4-Carboxyphenyl)-1H-1,2,3-triazol-4-yl)methyl)-4-oxo-3,4-dihydroquinazoline-2-carboxylic acid (6q)
Off-white solid, Rf (5% methanol in DCM): 0.16, anal (C19H13N5O5) calc. C 58.31 H 3.35 N 17.90, found: C 58.29 H 3.33 N 17.87. 1H-NMR (300 MHz, DMSO-d6) (δ, ppm): 13.24 (brs, 1H, OH), 12.65 (s, 1H, OH), 9.01 (s, 1H, Ar–H), 8.61 (s, 1H, CH), 8.17–8.02 (m, 4H, Ar–H), 7.95–7.82 (m, 1H, Ar–H), 7.72–7.68 (m, 1H, Ar–H), 7.58–7.53 (m, 1H, Ar–H), 5.39 (s, 2H, CH2). 13C-NMR (75 MHz, DMSO-d6) (δ, ppm): 166.81, 160.43, 148.41, 148.33, 144.53, 139.87, 134.91, 131.12, 127.72, 127.63, 126.56, 122.43, 122.15, 120.24, 116.90, 41.50. IR νmax(neat): 3064 ( CH), 1714 (C O, acid) and 1667 (C O, amide) cm−1. LC-MS: (m/z) [M − H]− 390.8.
3.3. X-ray crystallographic analysis
The structure of the compound 2 and 5c was unequivocally established by X-ray crystallographic analysis. Single crystals of 2 and 5c were obtained through the slow evaporation of their hexane–ethyl acetate solution. A suitable crystal was selected and mounted on a Kryoloop using paratone oil on Excalibur, Onyx, Ultra diffractometer. The crystal was kept at 100 K during data collection. Using Olex2,27 the structure was solved by direct method using ShelXS28 structure solution program and refined with the ShelXL29 refinement package using Least Squares minimisation.
3.4. Biology
3.4.1. In vitro anticandidal activity
All the synthesized compounds (5a–q and 6a–q) were screened for antifungal activities against C. albicans (ATCC 90028), C. glabrata (ATCC 90030), C. tropicalis (ATCC 750) using broth dilution technique according to standard protocol for antifungal assessment by NCCLS.30 Fluconazole (FLC) was used as positive control. Test compounds were dissolved in DMSO and further diluted in PBS to maintain appropriate concentration of DMSO to perform biological assays. Further, the desired concentration (1000 μg mL−1 to 7.8 μg mL−1) of test compounds were dispensed into 96 well plate and maintained in 100 μL Sabouraud Dextrose (SD) broth medium. The 100 μL of freshly prepared Candida spp. suspension containing approximately 2.5 × 103cells per mL were transferred into each well of 96 well plate (Tarson). The plate was incubated at 30 °C and constant stirring of 150 rpm. After 24 h, the optical density was measured at 580 nm using Thermo Scientific's Multiskan Go plate reader. The IC50 value was defined as the concentration of the test compound that causes 50% decrease in absorbance compared with that of the control (i.e. untreated cells). The IC50 values were calculated by plotting the graph between concentration (in 10log) and % inhibition of fungal cells. All the experiments were done in triplicate in separate time.
3.4.2. Hemolytic activity
Previously reported method was used to determine hemolytic activities of selected lead compounds on human red blood cells (hRBCs).31 Fluconazole was used as standard drug for comparison. 2 mL of human blood was obtained from healthy individual and collected in EDTA containing tubes. The sample was centrifuged at room temperature for 10 min and at 2000 rpm. Erythrocytes were harvested as pellet and washed thrice in phosphate buffer saline (PBS) solution. To the pellet, PBS was added to yield a 10% (v/v) erythrocytes/PBS suspension. The 10% suspension was then diluted 1 : 10 in PBS. From each suspension, 100 μL was added in triplicate to 100 μL of a different dilution series of test compounds in the same buffer in micro-centrifuged tubes. 1% Triton X-100 solution was used for 100% cell lyses. Complete set of tubes was incubated at 37 °C for 1 h which is followed by centrifugation for 10 min at 2000 rpm and at room temperature. Then 150 μL of supernatant fluid was transferred to a flat-bottomed microtiter plate (Tarson) and absorbance was measured at 450 nm using Thermo Scientific's Multiskan Go plate reader. The hemolysis percentage was calculated by following equation:% Hemolysis = [(A450 of test compound treated sample − A450 of buffer treated sample)/(A450 of 1% Triton X − A450 of buffer treated sample)] × 100where A450 is absorbance at 450 nm.
3.4.3. Cytotoxicity by MTT assay
Cytotoxicity of selected lead compounds was checked by MTT assay. MTT (3-(4,5-dimethyl-2-yl)-2,5-diphenyl tetrazolium bromide) was purchased from Sigma Aldrich. Dulbecco's modified Eagle's medium (DMEM), 0.25% trypsin, 0.02% EDTA mixture and Fetal bovine serum (FBS) were purchased from Gibco (Grand Island, NY). Human embryonic kidney (HEK-293) cell line was procured from National Centre for Cell Sciences (NCCS), Pune, India. The cells were cultured and maintained as a monolayer in DMEM supplemented with 10% FBS and antibiotics (100 units per mL penicillin and 100 μg mL−1 streptomycin) at 37 °C in humidified atmosphere of 5% CO2 in T-25 flasks. The cells were subcultured not more than twelve week or 30 passages. Cell count of approximate 8 × 103 cells per well were seeded in 96-well plate and set to incubate for 24 h before treatment. The cells were then treated with varying concentrations (10–200 μg mL−1) of the test compounds. After 24 h of incubation at 37 °C, the exhausted serum supplemented medium was removed and serum free media (50 μL) was added in each well. After that, 20 μL per well of MTT at a concentration of 5 mg mL−1 in PBS was added to each well and the plates were incubated for 4 h at 37 °C. Formazan crystals, the metabolized MTT product, were solubilized in DMSO (150 μL per well) and was quantified by reading the absorbance at 570 nm after incubation of 10 min on iMark Microplate Reader (Bio-Rad, Hercules, CA). All assays were performed in triplicate. Percent viability was taken as the relative absorbance of treated versus untreated control cells.
3.4.4. Growth curve studies
The C. albicans cells were freshly revived by subculture on the SD agar plate. A loop of inoculums was introduced into the SD broth and cells were grown for 16 h at 30 °C before use. Approximately 2 × 103 cells per mL was then inoculated into the freshly prepared 50 mL sterile SD medium. Different concentrations, equivalent to 2MIC, MIC, MIC/2, of compound 5n were added separately into the conical flasks containing inoculated medium and incubated at 30 °C and 160 rpm. Strain specific concentration of FLC was used as positive control viz. 31.25 μg mL−1 for C. albicans. At predetermined time periods (0, 2, 4, 6, 8, 10, 12, 14, 16, 18, 20, 22, and 24 h) after incubation with agitation at 37 °C, 1 mL aliquot to each sample was removed from the conical flask and growth was measured turbidometrically at 600 nm using Thermo Multiskan spectrophotometer. Optical density (O.D.) was recorded for each concentration against time (h).
3.4.5. Sterol quantitation method
The total intracellular sterols were extracted as reported earlier with slight modifications.22,23 50 mL of SD broth medium prepared in four conical flasks, sterilized and inoculated with freshly cultured cells of C. albicans ATCC 90028. All conical flasks were incubated at 30 °C and 160 rpm for 3 hours. Three conical flasks were then introduced with 2MIC, MIC and MIC/2 concentration while forth remain untreated. Again, all the conical flasks were incubated at 30 °C and 160 rpm for 16 h. After incubation, the cells were harvested by centrifugation at 3000 rpm for 3 min and weight. The pellet was treated with 25% alcoholic potassium hydroxide (KOH) solution followed by incubation at 85 °C for 1 h. After incubation, sterol was extracted by the addition of n-heptane : distilled water (1 : 3). The heptane layers were transferred in fresh test tube, diluted five-fold in 100% ethanol and scanned spectrophotometrically between 230 and 300 nm. The presence of ergosterol and the late sterol intermediate 24(28) dehydroergosterol (DHE) in the extracted sample resulted in a characteristic four peak curve. Both ergosterol and 24 (28) DHE absorb at 281.5 nm, whereas only 24(28) DHE shows an intense spectral absorption band at 230 nm. The absence of detectable ergosterol in the extract was indicated as a flat line. Ergosterol content was calculated as percentage wet weight of the cells by the following equation:% Ergosterol + % 24(28)DHE = [(A281.5/290) × F]/weight of pellethere, % 24(28)DHE = [(A230/518) × F]/pellet weight, A281.5 and A230 absorbance at 281.5 nm and 230 nm, respectively and F is the dilution factor in alcohol.
3.5. Docking studies
Molecular docking is a well-known computational method for drug discovery which is used to suggest the interaction between a ligand and a protein at the atomic level and predict to characterize the behavior of ligand in the binding site of target proteins. Molecular docking mockups were made on AutoDock 4.2 software.33 3D structure of the compound 5n was drawn in ChemDraw and the minimum energy conformation was calculated with Chem3D. The three-dimensional crystal structure of CYP51 (PDB ID: 5v5z) from the pathogen C. albicans was obtained from Protein Data Bank (https://www.rcsb.org/pdb). We used ADT tools to process the protein and ligand PDBs into ADT desired format (PDBQT). Protein processing includes removal of water molecules, the addition of hydrogen and charges to the protein and likewise ligand was processed for ADT format. Grid dimensions of X, Y, and Z covering the complete the CYP51 protein molecule was 46, 36 and 40 with 1 Å spacing. Dimensions of the centre grid box were −47.52, −14.83 and 22.93 in case of whole CYP51 molecule blind docking. Complex with the minimum binding energy and involving more number of interactions was chosen as the basis for further interface analysis. We mapped and visualized these interacting partners in PyMol.34
3.6. ADME prediction
A computational study of synthesized compounds 5a–q and 6a–q was performed for the prediction of ADME properties. In this study, molecular weight, number of hydrogen bond acceptor and donor functional groups, logarithm of the partition coefficient (log P), aqueous solubility, brain/blood partition coefficient, binding to human serum albumin and % human oral absorption were evaluated using QikProp version 5.1, Schrödinger software.35
4. Conclusion
In summary, a novel series of 1,2,3-triazole–quinazolinone conjugates (5a–q) and (6a–q) were prepared and evaluated for anticandidal activity. The in vitroanticandidal results show that all the synthesized compounds possess anticandidal activity to a certain extent. The compounds 5g, 5n and 6e show good activity against all the three strains of Candida, viz. C. albicans, C. glabrata and C. tropicalis. The compound 5n emerged as a most potent inhibitor among compounds (5a–q) and 6e among compounds (6a–q). The hemolysis and in vitro cytotoxicity results of compounds 5e, 5g, 5n, 6c, 6e, and 6n revealed non-toxic nature of these compounds. The sterol quantitation assay showed a significant decrease in ergosterol content which clearly indicates the interference of lead inhibitor 5n in sterol biosynthesis. Docking studies of 5n with lanosterol 14α-demethylase (CYP51) of C. albicans (PDB ID: 5v5z) support experimental data and aligned with the mechanism action of azoles. Further, in silico ADME prediction of synthesized compounds indicated that compounds have a potential to develop as good oral drug candidate for further biological studies.
5. Live subject statement
All experiments were performed in accordance with the Guidelines of “Clinical & Laboratory Standards Institute (CLSI)” using Departmental Instrumentation Facility (DIF) and experiments were approved by the Biosafety Committee at “Jamia Millia Islamia” University.
Conflicts of interest
All authors declare no conflict of interest.
Supplementary Material
Acknowledgments
Authors acknowledge the generous support from the Deanship of Scientific Research at King Saud University, Riyadh, Kingdom of Saudi Arabia, for funding this research group under grant No. RGP-150. M. M. Masood acknowledges the financial support from University Grants Commission (UGC), Govt. of India in the form of Teacher Fellow [F. No. 27-109/2014 (TF/NRCB)].
Electronic supplementary information (ESI) available. CCDC 1874110 and 1874111. For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/c8ra08426b
References
- Yao B. Ji H. Cao Y. Zhou Y. Zhu J. Lü J. Li Y. Chen J. Zheng C. Jiang Y. Rongmei-Liang A. Tang H H. J. Med. Chem. 2007;50:5293–5300. doi: 10.1021/JM0701167. doi: 10.1021/JM0701167. [DOI] [PubMed] [Google Scholar]
- Groll A. H. Lumb J. Future Microbiol. 2012;7:179–184. doi: 10.2217/fmb.11.154. doi: 10.2217/fmb.11.154. [DOI] [PubMed] [Google Scholar]
- Mendes Giannini M. J. S. Bernardi T. Scorzoni L. Fusco-Almeida A. M. Sardi J. C. O. J. Med. Microbiol. 2013;62:10–24. doi: 10.1099/jmm.0.045054-0. doi: 10.1099/jmm.0.045054-0. [DOI] [PubMed] [Google Scholar]
- Lu M. Li T. Wan J. Li X. Yuan L. Sun S. Int. J. Antimicrob. Agents. 2017;49:125–136. doi: 10.1016/j.ijantimicag.2016.10.021. doi: 10.1016/j.ijantimicag.2016.10.021. [DOI] [PubMed] [Google Scholar]
- Wang S. Wang Y. Liu W. Liu N. Zhang Y. Dong G. Liu Y. Li Z. He X. Miao Z. Yao J. Li J. Zhang W. Sheng C. ACS Med. Chem. Lett. 2014;5:506–511. doi: 10.1021/ml400492t. doi: 10.1021/ml400492t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X. Sun Z. Cao Y. Wang R. Cai T. Chu W. Hu W. Yang Y. J. Med. Chem. 2014;57:3687–3706. doi: 10.1021/jm4016284. doi: 10.1021/jm4016284. [DOI] [PubMed] [Google Scholar]
- Sheehan D. J. Hitchcock C. A. Sibley C. M. Clin. Microbiol. Rev. 1999;12:40–79. doi: 10.1128/CMR.12.1.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad A. Khan A. Manzoor N. Khan L. A. Microb. Pathog. 2010;48:35–41. doi: 10.1016/j.micpath.2009.10.001. doi: 10.1016/j.micpath.2009.10.001. [DOI] [PubMed] [Google Scholar]
- Bendaha H. Yu L. Touzani R. Souane R. Giaever G. Nislow C. Boone C. El Kadiri S. Brown G. W. Bellaoui M. Eur. J. Med. Chem. 2011;46:4117–4124. doi: 10.1016/j.ejmech.2011.06.012. doi: 10.1016/j.ejmech.2011.06.012. [DOI] [PubMed] [Google Scholar]
- Guillon R. Pagniez F. Giraud F. Crépin D. Picot C. Le Borgne M. Morio F. Duflos M. Logé C. Le Pape P. ChemMedChem. 2011;6:816–825. doi: 10.1002/cmdc.201000530. doi: 10.1002/cmdc.201000530. [DOI] [PubMed] [Google Scholar]
- Girmenia C. Finolezzi E. Clin. Invest. 2011;1:1577–1594. doi: 10.4155/cli.11.137. doi: 10.4155/cli.11.137. [DOI] [Google Scholar]
- Masood M. M. Pillalamarri V. K. Irfan M. Aneja B. Jairajpuri M. A. Zafaryab M. Rizvi M. M. A. Yadava U. Addlagatta A. Abid M. RSC Adv. 2015;5(43):34173–34183. doi: 10.1039/C5RA03354C. [DOI] [Google Scholar]
- Aneja B. Irfan M. Kapil C. Jairajpuri M. A. Maguire R. Kavanagh K. Rizvi M. M. A. Manzoor N. Azam A. Abid M. Org. Biomol. Chem. 2016;14(45):10599–10619. doi: 10.1039/C6OB01718E. [DOI] [PubMed] [Google Scholar]
- Masood M. M. Hasan P. Tabrez S. Ahmad M. B. Yadava U. Daniliuc C. G. Sonawane Y. A. Azam A. Rub A. Abid M. Bioorg. Med. Chem. Lett. 2017;9:1886–1891. doi: 10.1016/j.bmcl.2017.03.049. [DOI] [PubMed] [Google Scholar]
- Masood M. M. Irfan M. Alam S. Hasan P. Queen A. Shahid S. Zahid M. Azam A. Abid M. Lett. Drug Des. Discovery. 2018;15:1. doi: 10.2174/1570180815666180502120042. [DOI] [Google Scholar]
- Aneja B. Azam M. Alam S. Perwez A. Maguire R. Yadava U. Kavanagh K. Daniliuc C. G. Rizvi M. M. A. Haq Q. M. R. Abid M. ACS Omega. 2018;3(6):6912–6930. doi: 10.1021/acsomega.8b00582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aneja B. Irfan M. Hassan M. I. Prakash A. Yadava U. Daniliuc C. G. Zafaryab M. Rizvi M. M. A. Azam A. Abid M. J. Enzyme Inhib. Med. Chem. 2016;31(5):834–852. doi: 10.3109/14756366.2015.1058257. [DOI] [PubMed] [Google Scholar]
- Kelly S. L. David C. L. Andrew J. C. Baldwin B. C. Kelly D. E. Biochem. Biophys. Res. Commun. 1995;207(3):910–915. doi: 10.1006/bbrc.1995.1272. [DOI] [PubMed] [Google Scholar]
- Adams J. L. and Brian W. M., Therapeutic consequences of the inhibition of sterol metabolism, Pergamon Press, Oxford, England, vol. 2, 1990 [Google Scholar]
- Ji H. Zhang W. Zhou Y. Zhang M. Zhu J. Song Y. Lü J. Zhu J. J. Med. Chem. 2000;43(13):2493–2505. doi: 10.1021/jm990589g. [DOI] [PubMed] [Google Scholar]
- Váradi A. Horváth P. Kurtán T. Mándi A. Tóth G. Gergely A. Kökösi J. Tetrahedron. 2012;68:10365–10371. doi: 10.1016/j.tet.2012.09.086. doi: 10.1016/j.tet.2012.09.086. [DOI] [Google Scholar]
- Arthington-Skaggs A. B. Warnock D. W. Morrison C. J. Antimicrob. Agents Chemother. 2000;44(8):2081–2085. doi: 10.1128/AAC.44.8.2081-2085.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irfan M. Alam S. Manzoor N. Abid M. PLoS One. 2017;12(4):e0175710. doi: 10.1371/journal.pone.0175710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keniya M. V. Sabherwal M. Wilson R. K. Sagatova A. A. Tyndall J. D. A. Monk B. C. Antimicrob. Agents Chemother. 2018;62(11) doi: 10.1128/AAC.01134-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadekar P. K. Roychowdhury A. Kharkar P. S. Khedkar V. M. Arkile M. Manek H. Sarkar D. Sharma R. Vijayakumar V. Sarveswari S. Eur. J. Med. Chem. 2016;122:475–487. doi: 10.1016/j.ejmech.2016.07.001. doi: 10.1016/j.ejmech.2016.07.001. [DOI] [PubMed] [Google Scholar]
- Lipinski C. A. Lombardo F. Dominy B. W. Feeney P. J. Adv. Drug Delivery Rev. 1997;23:3–25. doi: 10.1016/S0169-409X(96)00423-1. doi: 10.1016/S0169-409X(96)00423-1. [DOI] [PubMed] [Google Scholar]
- Dolomanov O. V. Bourhis L. J. Gildea R. J. Howard J. A. K. Puschmann H. J. Appl. Crystallogr. 2009;42:339–341. doi: 10.1107/S0021889808042726. doi: 10.1107/S0021889808042726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourhis L. J. Dolomanov O. V. Gildea R. J. Howard J. A. K. Puschmann H. Acta Crystallogr., Sect. A: Found. Adv. 2015;71:59–75. doi: 10.1107/S2053273314022207. doi: 10.1107/S2053273314022207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldrick G. M. Acta Crystallogr., Sect. A: Found. Adv. 2015;71:3–8. doi: 10.1107/S2053273314026370. doi: 10.1107/S2053273314026370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikler M., Performance standards for antimicrobial susceptibility testing: seventeenth informational supplement, 2007 [Google Scholar]
- Irfan M. Aneja B. Yadava U. Khan S. I. Manzoor N. Daniliuc C. G. Abid M. Eur. J. Med. Chem. 2015;93:246–254. doi: 10.1016/j.ejmech.2015.02.007. [DOI] [PubMed] [Google Scholar]
- Lengauer T. Rarey M. Curr. Opin. Struct. Biol. 1996;6:402–406. doi: 10.1016/S0959-440X(96)80061-3. doi: 10.1016/S0959-440X(96)80061-3. [DOI] [PubMed] [Google Scholar]
- Trott O. Olson A. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano W., The PyMOL user's manual, DeLanoSci, San Carlos, CA, 2002 [Google Scholar]
- Schrödinger L., QikProp, version 5.1, New York, NY, 2017 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.