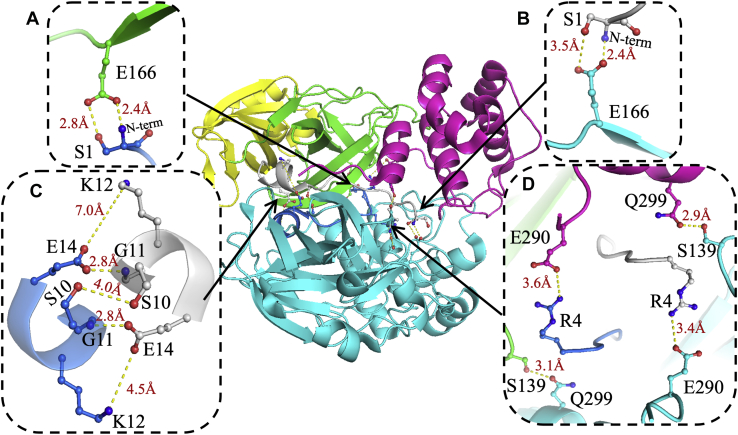
Figure 1.
Dimer interface interactions of SARS-CoV2 3CLpro. One monomer is shown in cyan, and the other monomer is colored by domain: N-finger in blue, domain I in yellow, domain II in green, and domain III in pink. Three sites of interactions were identified as important for 3CLpro dimer interface formation. A and B, at the first site, Glu166 in domain II interacts with the N terminus and side chain of Ser1 in the other monomer to form two points of contact between the two monomers. C, at the second site, the one-turn α-helix (residues 11–14) at the end of the N-finger of one monomer interacts with the same region of the other monomer to form a single contact point in the dimer interface. The side chains of the two Ser10 residues form intermolecular H-bonding interactions. Long-distance ionic interactions may occur between Lys12 of one monomer and Glu14 of the other monomer. D, the third site includes four residues. Arg4 of the N-finger interacts with Glu290 in domain III of the other monomer. In addition, Ser139 in domain II of one monomer interacts with Gln299 in domain III of the other monomer. The figure was generated using Protein Data Bank code 6WTM and PyMol (Schrodinger LLC) (31). 3CLpro, 3C-like protease; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.