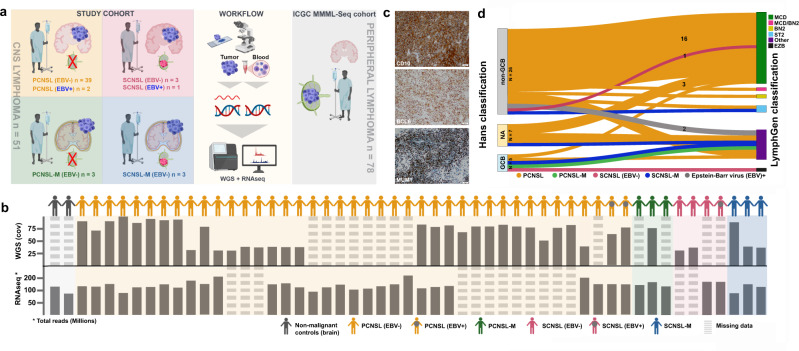
Fig. 1. Study design and multi-omic analysis of the CNSL cohort.
Panel a demonstrates the study design, study cohort along with sample size, and sequencing approach. The central nervous system lymphoma (CNSL) cohort consists of 51 primary and secondary lymphoma (PCNSL and SCNSL) patients. Whole-genome sequencing was performed using tumor tissue and matched peripheral blood samples. For RNA sequencing, we additionally analyzed normal controls (non-malignant brain tissue (frontal lobe)) and included data from peripheral lymphomas without CNS manifestation for validation (ICGC MMML-seq cohort). The lower panel b lists all CNS lymphoma derived from PCNSL or SCNSL patients as well as controls. The depth of coverage (cov) for whole-genome sequencing and total number of reads in RNA sequencing are given for each tumor sample. Whole-genome sequencing data were obtained from n = 38 PCNSL/SCNSL patients. RNA sequencing data was generated from n = 37 PCNSL/SCNSL patients and n = 2 normal controls. In 24 PCNSL/SCNSL cases, we obtained patient-matched whole-genome and RNA sequencing data. Striped bars indicate data sets missing for individual samples. The CNSL specimen were examined according to the Hans classification (CD10, BCL6, and MUM1 (c)). Additionally, we classified all n = 38 whole-genome sequencing CNSL samples according to the genetic DLBCL subtypes using the LymphGen algorithm as described by Wright et al., 2020 (d). The results are displayed in a Sankey plot. Images in panels a and b were partially created with BioRender.com.