Chemical structures, experimental and predicted pEC50 values of diarylpyridine derivatives acting as HIV-1 NNRTIs.
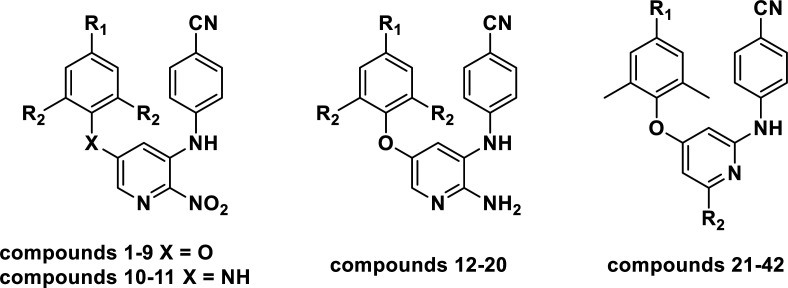
| ||||||||
|---|---|---|---|---|---|---|---|---|
| No. | R1 | R2 | EC50 (μM) | Actual pEC50 | CoMFA | CoMSIA | ||
| Predicted pEC50 | Residual | Predicted pEC50 | Residual | |||||
| 1 | CN | CH3 | 4.12 | 5.385 | 5.374 | 0.011 | 5.418 | −0.033 |
| 2a | CH3 | CH3 | 0.29 | 6.538 | 6.652 | −0.114 | 6.607 | −0.069 |
| 3b | CH3 | Br | 0.05 | 7.301 | 7.174 | 0.127 | 7.098 | 0.203 |
| 4 | H | CH3 | 2.03 | 5.693 | 5.680 | 0.013 | 5.707 | −0.014 |
| 5 | Cl | CH3 | 1.41 | 5.851 | 5.788 | 0.063 | 5.717 | 0.134 |
| 6 | Br | CH3 | 4.41 | 5.356 | 5.377 | −0.021 | 5.385 | −0.029 |
| 7 | Cl | Cl | 0.31 | 6.509 | 6.528 | −0.019 | 6.490 | 0.019 |
| 8 | Br | Br | 0.79 | 6.102 | 6.321 | −0.219 | 6.334 | −0.232 |
| 9 | F | F | 1.01 | 5.996 | 5.858 | 0.138 | 6.020 | −0.024 |
| 10 | H | CH3 | 1.06 | 5.975 | 6.061 | −0.086 | 6.000 | −0.025 |
| 11a,b | CH3 | CH3 | 0.04 | 7.398 | 7.409 | −0.011 | 6.831 | 0.567 |
| 12b | CN | CH3 | 0.07 | 7.155 | 7.088 | 0.067 | 7.176 | −0.021 |
| 13a | CH3 | CH3 | 0.14 | 6.854 | 6.978 | −0.124 | 6.950 | −0.096 |
| 14b | CH3 | Br | 0.07 | 7.155 | 7.249 | −0.094 | 7.191 | −0.036 |
| 15 | H | CH3 | 0.60 | 6.222 | 6.231 | −0.009 | 6.218 | 0.004 |
| 16 | H | OCH3 | 35.73 | 4.447 | 4.444 | 0.003 | 4.479 | −0.032 |
| 17 | Cl | CH3 | 0.37 | 6.432 | 6.310 | 0.122 | 6.336 | 0.096 |
| 18 | Br | CH3 | 0.84 | 6.076 | 6.206 | −0.130 | 6.153 | −0.077 |
| 19a | Cl | Cl | 0.26 | 6.585 | 6.522 | 0.063 | 6.362 | 0.223 |
| 20 | Br | Br | 0.50 | 6.301 | 6.245 | 0.056 | 6.228 | 0.073 |
| 21b | CH3 | Cl | 0.37 | 6.432 | 6.410 | 0.022 | 6.419 | 0.013 |
| 22b | CN | Cl | 0.84 | 6.076 | 6.125 | −0.049 | 6.108 | −0.032 |
| 23 | CH3 |
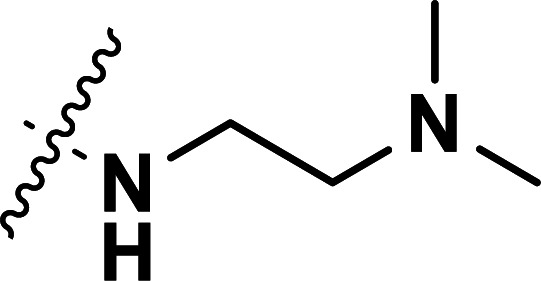
|
0.043 | 7.367 | 7.340 | 0.027 | 7.471 | −0.104 |
| 24 | CH3 |
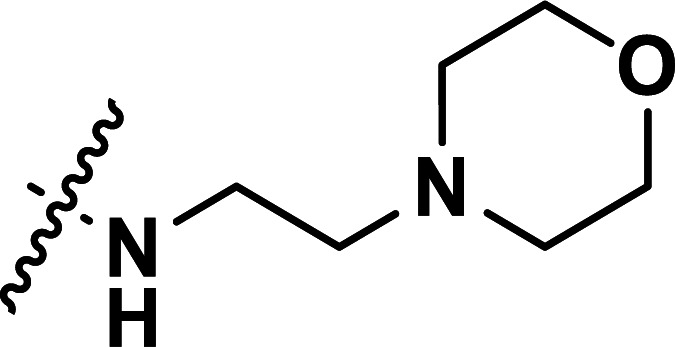
|
0.072 | 7.143 | 7.149 | −0.006 | 7.070 | 0.073 |
| 25 | CH3 |
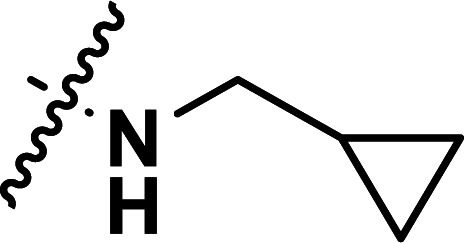
|
1.99 | 5.701 | 5.648 | 0.053 | 5.689 | 0.012 |
| 26a | CH3 |
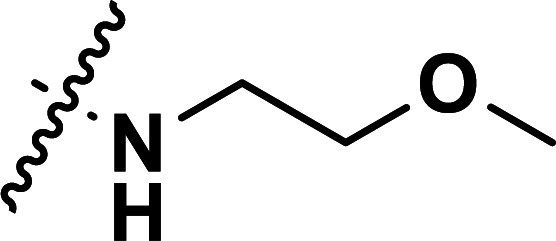
|
0.87 | 6.060 | 5.962 | 0.098 | 5.855 | 0.205 |
| 27 | CH3 |
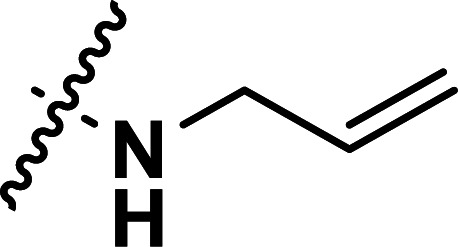
|
0.70 | 6.155 | 6.117 | 0.038 | 6.229 | −0.074 |
| 28b | CH3 |
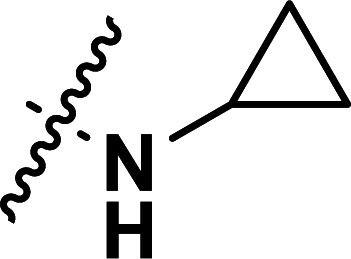
|
0.035 | 7.456 | 7.489 | −0.033 | 7.486 | −0.030 |
| 29 | CH3 |
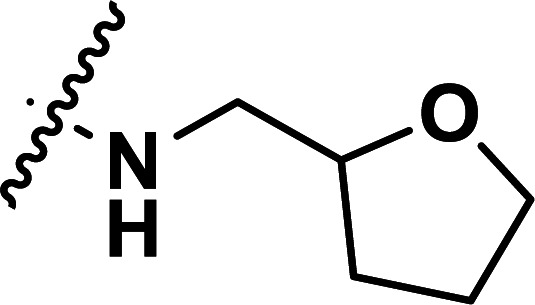
|
1.79 | 5.747 | 5.793 | −0.046 | 5.702 | 0.045 |
| 30a | CH3 |
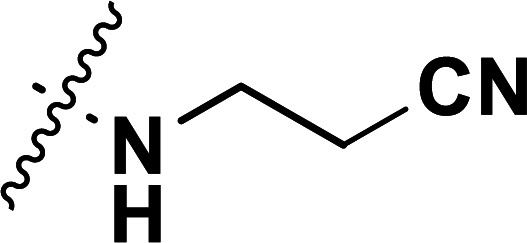
|
0.24 | 6.620 | 6.780 | −0.160 | 6.572 | 0.048 |
| 31 | CH3 |
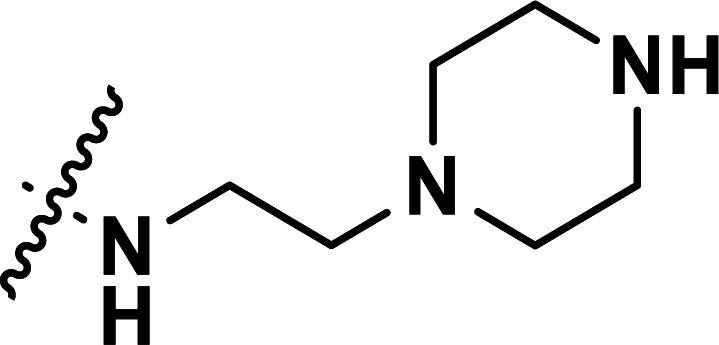
|
0.71 | 6.149 | 6.125 | 0.024 | 6.136 | 0.013 |
| 32 | CH3 |
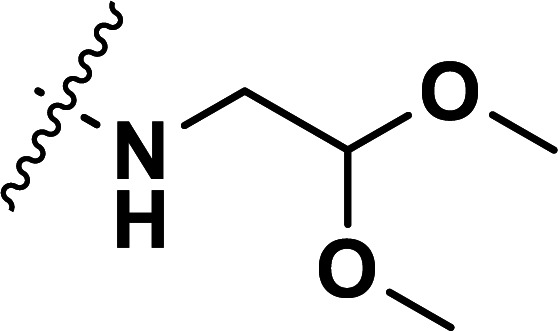
|
1.63 | 5.788 | 5.793 | −0.005 | 5.789 | −0.001 |
| 33b | CN |
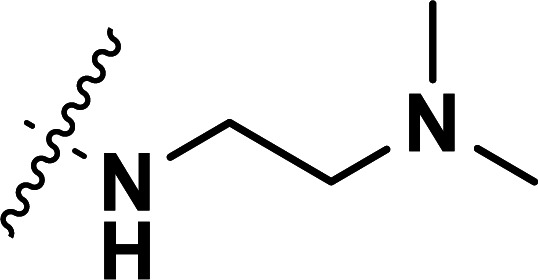
|
0.041 | 7.387 | 7.321 | 0.066 | 7.390 | −0.003 |
| 34b | CN |
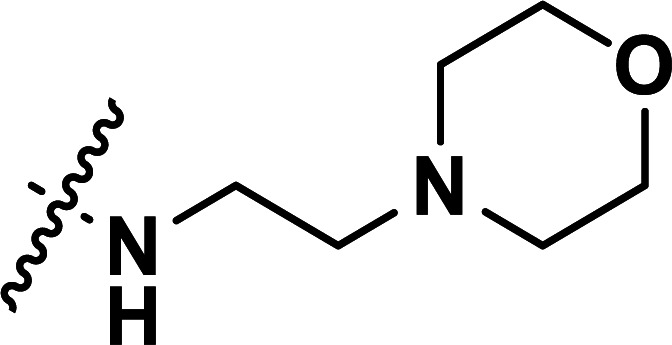
|
0.082 | 7.086 | 7.121 | −0.035 | 7.138 | −0.052 |
| 35 | CN |
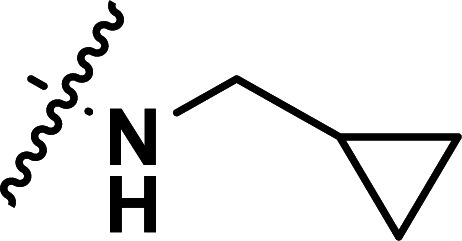
|
1.52 | 5.818 | 5.870 | −0.052 | 5.800 | 0.018 |
| 36a | CN |
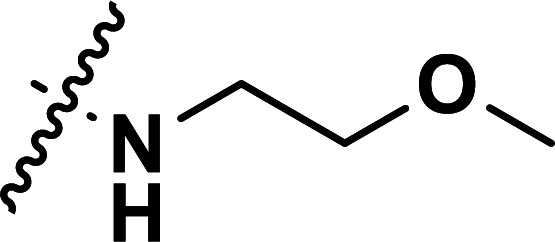
|
0.25 | 6.602 | 6.665 | −0.063 | 6.658 | −0.056 |
| 37 | CN |
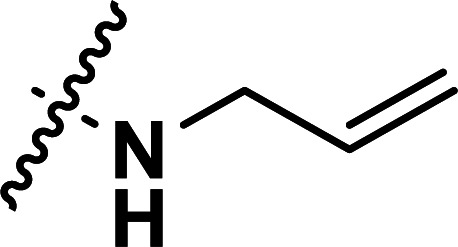
|
0.40 | 6.398 | 6.386 | 0.012 | 6.262 | 0.136 |
| 38 | CN |
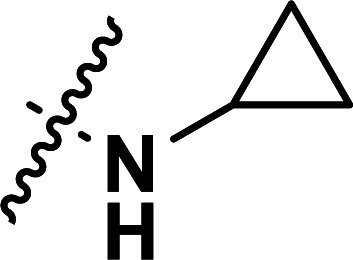
|
0.22 | 6.658 | 6.675 | −0.017 | 6.640 | 0.018 |
| 39 | CN |
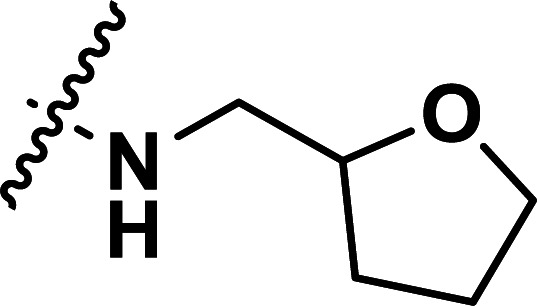
|
1.32 | 5.879 | 5.845 | 0.034 | 5.893 | −0.014 |
| 40b | CN |
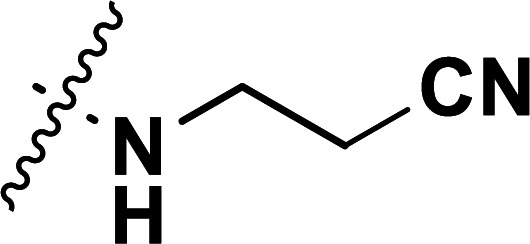
|
0.10 | 7.000 | 7.031 | −0.031 | 7.002 | −0.002 |
| 41 | CN |
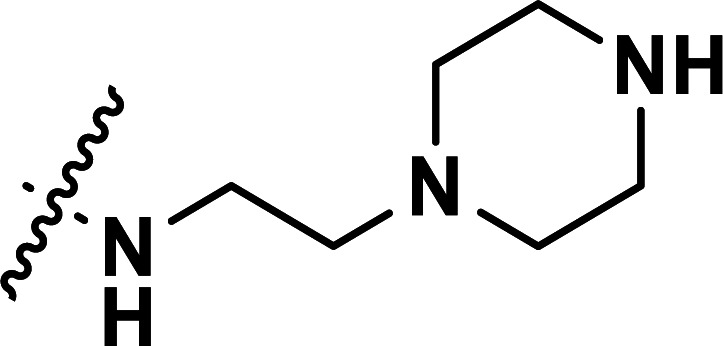
|
0.43 | 6.367 | 6.357 | 0.010 | 6.383 | −0.016 |
| 42 | CN |
|
0.84 | 6.076 | 6.109 | −0.033 | 6.079 | −0.003 |
Test set compounds of 3D-QSAR models.
Compounds used for pharmacophore models.
